Abstract
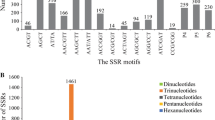
The increasing availability of expressed sequence tags (ESTs) in wheat (Triticum aestivum) and related cereals provides a valuable resource of non-anonymous DNA molecular markers. We examined 170,746 wheat ESTs from the public (International Triticeae EST Cooperative) and Génoplante databases, previously clustered in contigs, for the presence of di- to hexanucleotide simple sequence repeats (SSRs). Analysis of 46,510 contigs identified 3,530 SSRs, which represented 7.5% of the total number of contigs. Only 74% of the sequences allowed primer pairs to be designed, 70% led to an amplification product, mainly of a high quality (68%), and 53% exhibited polymorphism for at least one cultivar among the eight tested. Even though dinucleotide SSRs were less represented than trinucleotide SSRs (15.5% versus 66.5%, respectively), the former showed a much higher polymorphism level (83% versus 46%). The effect of the number and type of repeats is also discussed. The development of new EST-SSRs markers will have important implications for the genetic analysis and exploitation of the genetic resources of wheat and related species and will provide a more direct estimate of functional diversity.


Similar content being viewed by others
References
Benson G (1999) tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primer improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31:24–28
Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104:399–407
Feillet P (1965) Contribution à l’étude des proteines de blé. Influence des facteurs génétiques, agronomiques et technologiques. Ann Technol Agric 14:95
Gupta PK, Balyan HS, Edwards KJ, Isaac P, Korzun V, Röder M, Gautier M-F, Joudrier P, Schlatter AR, Dubcovsky J, De la Pena RC, Khairallah M, Penner G, Hayden MJ, Sharp P, Keller B, Wang RCC, Hardouin JP, Jack P, Leroy P (2002) Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat. Theor Appl Genet 105:413–422
Guyomarc’h H, Sourdille P, Charmet G, Edwards KJ, Bernard M (2002) Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor Appl Genet 104:1164–1172
Holton TA, Christopher JT, McClure L, Harker N, Henry RJ (2002) Identification and mapping of polymorphic SSR markers from expressed gene sequences of barley and wheat. Mol Breed 9:63–71
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequences repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Lagercrantz U, Ellegren H, Anderson L (1993) The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res 21:1111–1115
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Leroy P, Nègre S, Tixier MH, Perretant MR, Sourdille P, Gay G, Bernard M, Coville JL, Quétier F, Nelson C, Sorrells M, Marino CL, Hart G, Friebe B, Gill BS, Röder M (1997) A genetic reference map for the bread wheat genome, Triticum aestivum L. em. Thell. In: McGuire PE, Qualset CO (eds) Progress in genome mapping of wheat and related species. Joint Proc 5th and 6th Public Workshops Int Triticae Mapping Initiative. Report No. 18, University of California, Davis, pp 134–140
Lewin B (1994) Genes V. Oxford University Press, New York
Liu ZW, Biyashev RM, Maroof MAS (1996) Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor Appl Genet 93:869–876
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol 35:89–99
Morgante M, Olivieri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3:175–182
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with non repetitive DNA in plant genomes. Nat Genet 30:194–200
Murray EE, Lotzer J, Eberle M (1989) Codon usage in plant genes. Nucleic Acids Res 17:477–498
Ramsay L, Macaulay M, Cardle L, Morgante M, Degli Ivanissevich S, Maestri E, Powell W, Waugh R (1999) Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J 17:415–425
Ramsay L, Macaulay M, degli Ivannissevich S, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R (2000) A simple sequence repeat-based linkage map of barley. Genetics 156:1997–2005
Röder MS, Plaschke J, König SU, Börner A, Sorrells ME, Tanksley SD, Ganal MW (1995) Abundance, variability and chromosomal location of microsatellites in wheat. Mol Gen Genet 246:327–333
Röder MS, Korsun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of the wheat genome. Genetics 149:2007–2023
Scott KD, Eggler P, Seaton P, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Senior ML, Chin ECL, Lee M, Smith JSC (1996) Simple sequence repeat markers developed from maize found in the GenBank database: map construction. Crop Sci 36:1676–1683
Sourdille P, Tavaud M, Charmet G, Bernard M (2001a) Transferability of wheat microsatellites to diploid Triticeae species carrying the A, B, and D genome. Theor Appl Genet 103:346–352
Sourdille P, Guyomarc’h H, Baron C, Gandon B, Chiquet V, Artiguenave F, Edwards K, Foisset N, Dufour P, Bernard M (2001b) Improvement of the genetic maps of wheat using new microsatellite markers. In: Proc 9th Plant Anim Genome. Final abstract guide. Applied Biosystems (sponsors), Foster City, pp 167, 425
Temnykh S, Park WD, Ayers N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (1999) Mapping and genome organization of microsatellites in rice Oryza sativa. Theor Appl Genet 100:698–712
Tixier MH, Sourdille P, Röder M, Leroy P, Bernard M (1997) Detection of wheat microsatellites using a non-radioactive silver nitrate staining method. J Genet Breed 51:175–177
Tixier MH, Sourdille P, Charmet G, Gay G, Jaby C, Cadalen T, Bernard S, Nicolas P, Bernard M (1998) Detection of QTLs for crossability in wheat using a double-haploid population. Theor Appl Genet 97:1076–1082
Van Deynze AE, Dubcovsky J, Gill KS, Nelson JC, Sorrels ME, Dvorak J, Gill BS, Lagudah ES, McCouch SR, Appels R (1995) Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oat. Genome 38:45–59
Acknowledgements
The authors gratefully acknowledge B. Charef for excellent technical assistance and G. Gay and A. Loussert for growing the plants. S. Reader and B. Gill are acknowledged for furnishing the aneuploid and deletion lines. This work was supported by Génoplante, the French joint program in plant genomics. All of the experiments described herein comply with the current laws of France.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.W. Snape
Rights and permissions
About this article
Cite this article
Nicot, N., Chiquet, V., Gandon, B. et al. Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs). Theor Appl Genet 109, 800–805 (2004). https://doi.org/10.1007/s00122-004-1685-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1685-x