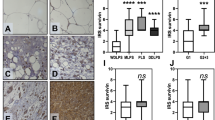
Abstract
Liposarcoma (LPS) is the most common soft tissue sarcoma and accounts for approximately 20 % of all adult sarcomas. Current treatment modalities (surgery, chemotherapy, and radiotherapy) all have limitations; therefore, molecularly driven studies are needed to improve the identification and increased understanding of genetic and epigenetic deregulations in LPS if we are to successfully target specific tumorigenic drivers. It can be anticipated that such biology-driven therapeutics will improve treatments by selectively deleting cancer cells while sparing normal tissues. This review will focus on several therapeutically actionable molecular markers identified in well-differentiated LPS and dedifferentiated LPS, highlighting their potential clinical applicability.

Similar content being viewed by others
References
American Cancer Society (2015) Cancer facts & figures 2015. American Cancer Society, Atlanta
Fletcher CDM, Bridge J, Hogendoorn P et al (2013) World Health Organization Classification of tumours pathology and genetics of tumours of soft tissue and bone, 4th edn. IARC Press, Lyon
Doyle LA (2014) Sarcoma classification: an update based on the 2013 World Health Organization Classification of tumors of soft tissue and bone. Cancer 120:1763–1774. doi:10.1002/cncr.28657
Dalal KM, Kattan MW, Antonescu CR et al (2006) Subtype specific prognostic nomogram for patients with primary liposarcoma of the retroperitoneum, extremity, or trunk. Ann Surg 244:381–391. doi:10.1097/01.sla.0000234795.98607.00
Bonvalot S, Rivoire M, Castaing M et al (2009) Primary retroperitoneal sarcomas: a multivariate analysis of surgical factors associated with local control. J Clin Oncol 27:31–37. doi:10.1200/JCO.2008.18.0802
Dei Tos AP (2000) Liposarcoma: new entities and evolving concepts. Ann Diagn Pathol 4:252–266. doi:10.1053/adpa.2000.8133
Lahat G, Anaya DA, Wang X et al (2008) Resectable well-differentiated versus dedifferentiated liposarcomas: two different diseases possibly requiring different treatment approaches. Ann Surg Oncol 15:1585–1593. doi:10.1245/s10434-007-9805-x
Azumi N, Curtis J, Kempson RL, Hendrickson MR (1987) Atypical and malignant neoplasms showing lipomatous differentiation. A study of 111 cases. Am J Surg Pathol 11:161–183
Keung EZ, Hornick JL, Bertagnolli MM et al (2014) Predictors of outcomes in patients with primary retroperitoneal dedifferentiated liposarcoma undergoing surgery. J Am Coll Surg 218:206–217. doi:10.1016/j.jamcollsurg.2013.10.009
Fabre-Guillevin E, Coindre J-M, de Saint Aubain Somerhausen N et al (2006) Retroperitoneal liposarcomas: follow-up analysis of dedifferentiation after clinicopathologic reexamination of 86 liposarcomas and malignant fibrous histiocytomas. Cancer 106:2725–2733
Singer S, Antonescu CR, Riedel E, Brennan MF (2003) Histologic subtype and margin of resection predict pattern of recurrence and survival for retroperitoneal liposarcoma. Ann Surg 238:358–70. doi:10.1097/01.sla.0000086542.11899.38 (Discussion 370–371)
Evans HL (1979) Liposarcoma: a study of 55 cases with a reassessment of its classification. Am J Surg Pathol 3:507–523
Ghadimi MP, Al-Zaid T, Madewell J et al (2011) Diagnosis, management, and outcome of patients with dedifferentiated liposarcoma systemic metastasis. Ann Surg Oncol 18:3762–3770. doi:10.1245/s10434-011-1794-0
Evans HL, Khurana KK, Kemp BL, Ayala AG (1994) Heterologous elements in the dedifferentiated component of dedifferentiated liposarcoma. Am J Surg Pathol 18:1150–1157
Henricks WH, Chu YC, Goldblum JR, Weiss SW (1997) Dedifferentiated liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol 21:271–281
Tseng WW, Madewell JE, Wei W et al (2014) Locoregional disease patterns in well-differentiated and dedifferentiated retroperitoneal liposarcoma: implications for the extent of resection? Ann Surg Oncol 21:2136–2143. doi:10.1245/s10434-014-3643-4
Thway K, Jones RL, Noujaim J et al (2016) Dedifferentiated Liposarcoma: updates on morphology, genetics, and therapeutic strategies. Adv Anat Pathol 23:30–40. doi:10.1097/PAP.0000000000000101
Evans HL (1979) Liposarcoma a study of 55 cases with a reassessment of its classification. Am J Surg Pathol 3(6):507–523
Henricks WH, Chu YC, Goldblum JR, Weiss SW (1997) Dedifferentiated Liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Clin Pathol 21:271–281
Evans HL, Khurana KK, Kemp BL, Ayala AG (1994) Heterologous elements in the dedifferentiated components of dedifferentiated liposarcoma. Am J Clin Pathol 18:1077–1182
Mariño-Enríquez A, Fletcher CDM, Dal Cin P, Hornick JL (2010) Dedifferentiated liposarcoma with “homologous” lipoblastic (pleomorphic liposarcoma-like) differentiation: clinicopathologic and molecular analysis of a series suggesting revised diagnostic criteria. Am J Surg Pathol 34:1122–1131. doi:10.1097/PAS.0b013e3181e5dc49
Boland JM, Weiss SW, Oliveira AM et al (2010) Liposarcomas with mixed well-differentiated and pleomorphic features: a clinicopathologic study of 12 cases. Am J Surg Pathol 34:837–843. doi:10.1097/PAS.0b013e3181dbf2f7
Mandahl N, Höglund M, Mertens F et al (1994) Cytogenetic aberrations in 188 benign and borderline adipose tissue tumors. Genes Chromosomes Cancer 9:207–215
Szymanska J, Virolainen M, Tarkkanen M et al (1997) Overrepresentation of 1q21–23 and 12q13–21 in lipoma-like liposarcomas but not in benign lipomas: a comparative genomic hybridization study. Cancer Genet Cytogenet 99:14–18
Crago AM, Singer S (2011) Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol 23:373–378. doi:10.1097/CCO.0b013e32834796e6
Italiano A, Bianchini L, Gjernes E et al (2009) Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin Cancer Res 15:5696–5703. doi:10.1158/1078-0432.CCR-08-3185
Pilotti S, Della Torre G, Lavarino C et al (1998) Molecular abnormalities in liposarcoma: role of MDM2 and CDK4-containing amplicons at 12q13–22. J Pathol 185:188–190. doi:10.1002/(SICI)1096-9896(199806)185:2<188:AID-PATH53>3.0.CO;2-2
Dei Tos AP, Doglioni C, Piccinin S et al (2000) Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol 190:531–536
Segura-Sánchez J, González-Cámpora R, Pareja-Megia MJ et al (2006) Chromosome-12 copy number alterations and MDM2, CDK4 and TP53 expression in soft tissue liposarcoma. Anticancer Res 26:4937–4942
Shimoji T, Kanda H, Kitagawa T et al (2004) Clinico-molecular study of dedifferentiation in well-differentiated liposarcoma. Biochem Biophys Res Commun 314:1133–1140
Matushansky I, Hernando E, Socci ND et al (2008) A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am J Pathol 172:1069–1080. doi:10.2353/ajpath.2008.070284
Kollár A, Benson C (2014) Current management options for liposarcoma and challenges for the future. Expert Rev Anticancer Ther 14:297–306. doi:10.1586/14737140.2014.869173
Bill KLJ, Garnett J, Meaux I et al (2016) SAR405838: a novel and potent inhibitor of the MDM2:p53 axis for the treatment of dedifferentiated liposarcoma. Clin Cancer Res 22:1150–1160. doi:10.1158/1078-0432.CCR-15-1522
Zhang Y-X, Sicinska E, Czaplinski JT et al (2014) Antiproliferative Effects of CDK4/6 inhibition in CDK4-amplified human liposarcoma in vitro and in vivo. Mol Cancer Ther 13:2184–2193. doi:10.1158/1535-7163.MCT-14-0387
Sandberg AA (2004) Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: liposarcoma. Cancer Genet Cytogenet 155:1–24. doi:10.1016/j.cancergencyto.2004.08.005
Wang X, Asmann YW, Erickson-johnson MR et al (2011) High-resolution genomic mapping reveals consistent amplification of the fibroblast growth factor receptor substrate 2 gene in well-differentiated and dedifferentiated liposarcoma. Genes Chromosomes Cancer 858:849–858. doi:10.1002/gcc
Haluska FG, Huebner K, Isobe M et al (1988) Localization of the human JUN protooncogene to chromosome region 1p31-32. Proc Natl Acad Sci USA 85:2215–2218
Mariani O, Brennetot C, Coindre J-M et al (2007) JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 11:361–374. doi:10.1016/j.ccr.2007.02.007
Snyder EL, Sandstrom DJ, Law K et al (2009) c-Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol 218:292–300. doi:10.1002/path.2564
Chibon F, Mariani O, Derré J et al (2004) ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14–q15 and 6q23 amplifications. Genes Chromosomes Cancer 40:32–37. doi:10.1002/gcc.20012
Schmidt H, Bartel F, Kappler M et al (2005) Gains of 13q are correlated with a poor prognosis in liposarcoma. Mod Pathol 18:638–644. doi:10.1038/modpathol.3800326
Wisdom R, Johnson RS, Moore C (1999) c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J 18:188–197. doi:10.1093/emboj/18.1.188
Cortner J, Vande Woude GF, Rong S (1995) The Met-HGF/SF autocrine signaling mechanism is involved in sarcomagenesis. EXS 74:89–121
Rong S, Jeffers M, Resau JH et al (1993) Met Expression and sarcoma tumorigenicity. Cancer Res 53:5355–5360
Ferracini R, Olivero M, Di Renzo MF et al (1996) Retrogenic expression of the MET proto-oncogene correlates with the invasive phenotype of human rhabdomyosarcomas. Oncogene 12:1697–1705
Rao UN, Sonmez-Alpan E, Michalopoulos GK (1997) Hepatocyte growth factor and c-MET in benign and malignant peripheral nerve sheath tumors. Hum Pathol 28:1066–1070
Torres KE, Zhu Q-S, Bill K et al (2011) Activated MET is a molecular prognosticator and potential therapeutic target for malignant peripheral nerve sheath tumors. Clin Cancer Res 17:3943–3955. doi:10.1158/1078-0432.CCR-11-0193
Que W, Chen J (2011) Knockdown of c-Met inhibits cell proliferation and invasion and increases chemosensitivity to doxorubicin in human multiple myeloma U266 cells in vitro. Mol Med Rep 4:343–349. doi:10.3892/mmr.2011.426
Peng T, Zhang P, Liu J et al (2011) An experimental model for the study of well-differentiated and dedifferentiated liposarcoma; deregulation of targetable tyrosine kinase receptors. Lab Invest 91:392–403. doi:10.1038/labinvest.2010.185
Bill KLJ, Garnett J, Ma X et al (2015) The hepatocyte growth factor receptor as a potential therapeutic target for dedifferentiated liposarcoma. Lab Investig 95:951–961. doi:10.1038/labinvest.2015.62
Bill KLJ, Pollock RE, Chen J (2015) Sensitivity to doxorubicin and MDM2 inhibitors correlate with MDM2 levels in dedifferentiated liposarcoma (abstract). In: 20th Annual meeting Connective Tissue Oncology Society
Ugras S, Brill E, Jacobsen A et al (2011) Small RNA sequencing and functional characterization reveals microRNA-143 tumor suppressor activity in liposarcoma. Cancer Res 71:5659–5669. doi:10.1158/0008-5472
Gits CMM, Van Kuijk PF, Jonkers MBE et al (2014) MicroRNA expression profiles distinguish liposarcoma subtypes and implicate miR-145 and miR-451 as tumor suppressors. Int J Cancer 135:348–361. doi:10.1002/ijc.28694
Lee DH, Amanat S, Goff C et al (2013) Overexpression of miR-26a-2 in human liposarcoma is correlated with poor patient survival. Oncogenesis 2:e47. doi:10.1038/oncsis.2013.10
Zhang P, Bill K, Liu J et al (2012) MiR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhances β-catenin signaling. Cancer Res 72:1751–1762. doi:10.1158/0008-5472.CAN-11-3027
Valadi H, Ekström K, Bossios A et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi:10.1038/ncb1596
Kinet V, Halkein J, Dirkx E, De Windt LJ (2013) Cardiovascular extracellular microRNAs: emerging diagnostic markers and mechanisms of cell-to-cell RNA communication. Front Genet 4:214. doi:10.3389/fgene.2013.00214
Fabbri M, Paone A, Calore F et al (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. PNAS 109:2110–2116. doi:10.1073/pnas.1209414109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1209414109
Strimbu K, Tavel JA (2010) What are biomarkers? Curr Opin HIV AIDS 5:463–466. doi:10.1097/COH.0b013e32833ed177
Panotopoulos J, Posch F, Alici B et al (2015) Hemoglobin, alkalic phosphatase, and C-reactive protein predict the outcome in patients with liposarcoma. J Orthop Res 33:765–770. doi:10.1002/jor.22827
Panotopoulos J, Posch F, Funovics PT et al (2015) Elevated serum creatinine and low albumin are associated with poor outcomes in patients with liposarcoma. J Orthop Res. doi:10.1002/jor.23002
Ballman KV (2015) Biomarker: predictive or prognostic? doi:10.1200/JCO.2015.63.3651
Schwarzenbach H, Nishida N, Calin GA, Pantel K (2014) Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 11:145–156. doi:10.1038/nrclinonc.2014.5
Subramanian S, Lui WO, Lee CH et al (2008) MicroRNA expression signature of human sarcomas. Oncogene 27:2015–2026. doi:10.1038/sj.onc.1210836
Sarver AL, Phalak R, Thayanithy V, Subramanian S (2010) S-MED: sarcoma microRNA expression database. Lab Investig 90:753–761. doi:10.1038/labinvest.2010.53
Renner M, Czwan E, Hartmann W et al (2012) MicroRNA profiling of primary high-grade soft tissue sarcomas. Genes Chromosomes Cancer 51:982–996. doi:10.1002/gcc.21980
Zhou Y, Zhang Y, Huang Y et al (2014) Liposarcoma miRNA signatures identified from genome-wide miRNA expression profiling. Future Oncol 10:1373–1386. doi:10.2217/fon.14.90
Taylor BS, DeCarolis PL, Angeles CV et al (2011) Frequent alterations and epigenetic silencing of differentiation pathway genes in structurally rearranged liposarcomas. Cancer Discov 1:587–597. doi:10.1158/2159-8290.CD-11-0181
Pichler M, Calin GA (2015) MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer 113:1–5. doi:10.1038/bjc.2015.253
Redis RS, Calin S, Yang Y et al (2012) Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther 136:169–174. doi:10.1016/j.pharmthera.2012.08.003
Taylor DD, Gercel-Taylor C (2013) The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet 4:1–12. doi:10.3389/fgene.2013.00142
Ma R, Jiang T, Kang X (2012) Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res 31:38. doi:10.1186/1756-9966-31-38
Miyachi M, Tsuchiya K, Yoshida H et al (2010) Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun 400:89–93. doi:10.1016/j.bbrc.2010.08.015
Weng Y, Chen Y, Chen J et al (2013) Identification of serum microRNAs in genome-wide serum microRNA expression profiles as novel noninvasive biomarkers for malignant peripheral nerve sheath tumor diagnosis. Med Oncol 30:531–536. doi:10.1007/s12032-013-0531-x
Fricke A, Ullrich PV, Heinz J et al (2015) Identification of a blood-borne miRNA signature of synovial sarcoma. Mol Cancer 14:151. doi:10.1186/s12943-015-0424-z
Acknowledgments
The authors would like to acknowledge Dr. Julie Bridge from the Department of Pathology, University of Nebraska who graciously provided the Fluorescence In Situ Hybridization (FISH) image.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This review was supported by the NCI U54CA168512 to REP.
Additional information
K. L. J. Bill and L. Casadei have contributed equally.
Rights and permissions
About this article
Cite this article
Bill, K.L.J., Casadei, L., Prudner, B.C. et al. Liposarcoma: molecular targets and therapeutic implications. Cell. Mol. Life Sci. 73, 3711–3718 (2016). https://doi.org/10.1007/s00018-016-2266-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2266-2