Abstract
Several dietary factors and their genetic modifiers play a role in neurological disease and affect the human brain. The structural and functional integrity of the living brain can be assessed using neuroimaging, enabling large-scale epidemiological studies to identify factors that help or harm the brain. Iron is one nutritional factor that comes entirely from our diet, and its storage and transport in the body are under strong genetic control. In this review, we discuss how neuroimaging can help to identify associations between brain integrity, genetic variations, and dietary factors such as iron. We also review iron’s essential role in cognition, and we note some challenges and confounds involved in interpreting links between diet and brain health. Finally, we outline some recent discoveries regarding the genetics of iron and its effects on the brain, suggesting the promise of neuroimaging in revealing how dietary factors affect the brain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human brain changes dynamically throughout life, and profound changes occur from childhood to old age, and, in particular, with the progression of disease. The structural integrity of the living brain may be evaluated with brain scans obtained through a range of neuroimaging techniques. These brain scans include, among others, high-resolution magnetic resonance imaging (MRI) and diffusion-based MRI. Standard anatomical MRI has been the mainstay of clinical neuroradiology for over two decades, and has helped reveal signs of brain aging, such as cortical atrophy, vascular changes, and changes over time in the gray and white matter. More recently, “diffusion-based” MRI has been embraced by neuroscientists and clinical researchers alike, as it can assess microstructural properties of the white matter fibers and the breadth of their connections. This technology is offering new insights into how the brain is organized, how it changes as we age, and what factors affect the changing brain.
Clearly, a wide range of neuroanatomical variation is expected in patients diagnosed with neurological or psychiatric illness. However, even in groups of healthy individuals, brain structure and integrity vary widely. For many measures of human brain morphometry and integrity, around half of the normal variance is due to the genetic variations among us. Even so, environmental factors also affect the brain—both structurally and functionally. Among these factors are education, diet, and stress.
One of the most basic—and to some extent controllable—influences on brain structure and function is a person’s diet and nutritional intake. In addition to proteins (and certain fats, vitamins, and other important minerals such as zinc, copper, iodine, selenium), iron is a nutrient with profound effects on the brain throughout life [1]. Iron’s effects on the brain have been a topic of interest for many years. Iron is critical for healthy brain development, and abnormal iron accumulation in the brain can promote neurodegenerative diseases [2]. In the past few decades, medical and public opinion has changed from favoring iron supplementation in healthy people as they age, to being more circumspect about its potential negative consequences [3].
Many nutrients, including iron, are not metabolized, transported, or processed identically in all individuals. They are regulated by a variety of genes, proteins, and their interactions. In most cases, iron enters the body solely through our diet, and exists as heme and non-heme iron in the body. Specific genes and regulatory proteins are involved in iron metabolism and are responsible for storing iron locally (ferritin) and transporting it (transferrin) throughout the body. The uptake and storage of iron in the brain differs across the brain structures and the distribution patterns also change with aging. It is perhaps not surprising then that genes involved in iron processing and transport are critical in shaping the human brain. However, the specific mechanisms of these effects are only just beginning to be understood.
Genetic diversity and the brain
In recent years, the field of “imaging genetics” [4] has matured significantly. Imaging genetics aims to correlate brain imaging measures from large groups of people to commonly carried variants in their DNA, in an effort to understand how genetic variation affects the brain and our risk for disease. Initial studies have offered a deeper understanding of how “disease risk” genes may affect the brain, and how they affect brain function and behavior [5].
Carriers of certain common genetic variants—or single-nucleotide polymorphisms (SNPs)—tend to have characteristic differences in brain structure that can be identified, at least at the group level, on MRI scans. These differences in brain integrity suggest how these genes may act to change neural circuitry and neural pathways, affecting a person’s aggregate risk for diseases such as Alzheimer’s disease, schizophrenia, and even autism. Some genetic variants are very common—and have substantial effects on the brain. These include commonly carried variants in the Alzheimer’s disease risk genes, ApoE [6], CLU [7], and MTHFR [8]. Brain variations have also been associated with carrying risk genes for psychiatric illness such as DISC1 [9] and neuregulin [10, 11], and risk genes for developmental disorders such as autism, including CNTNAP2 and MET [12, 13], among others. The flood of brain imaging and genetic data, available now on a large scale, has also made it possible to search the entire genome to identify new genetic variants that affect the volume of distinct parts of the brain [14–17].
In this review, we discuss the role of population-based neuroimaging studies in linking iron and other dietary factors to variations in brain integrity. We focus on iron, but briefly review other neuroimaging studies of diet and the brain. We also point out confounds and caveats to be aware of when interpreting these studies. Despite possible caveates, there has been progress in understanding molecular pathways that mediate dietary effects on the brain. Several genetic variants are associated with differences in iron metabolism, including its storage and transport; in fact, the most common genetic disorder in the world—hemochromatosis—is a disorder of iron metabolism, and affects 1 % of the population in some countries (e.g., Ireland). We describe current evidence relating iron levels to cognitive function, and new connections between brain integrity and genes that affect iron transport [18]. As brain databases expand to include ever-larger populations, neuroimaging can help to identify dietary factors and related genetic variants that consistently affect the brain, sometimes revealing mechanisms at the molecular level that mediate these effects.
Brain imaging and diet
Magnetic resonance imaging (MRI) and its extensions allow us to observe, measure, and quantify neuroanatomical structures in a non-invasive fashion. When large numbers of individuals are scanned, it is possible to study patterns of brain abnormalities due to disease, and localize subtle anatomical differences that relate to variations in cognitive abilities, or even to single base pair variations in the genome, as we noted earlier [14]. Structures of interest (such as the volume of the hippocampus—a key structure for learning and memory) can be measured, based on individual brain scans. The relative size and integrity of anatomical structures can be compared across entire populations to identify factors that affect them, adversely or positively. Additionally, brain mapping studies allow, through careful co-registration, each individual subject’s brain scan to be mapped into a common 3D coordinate space, enabling inter-subject or inter-group comparisons. In this sense, subjects’ brain data can be compared at all 3D points—or voxels—of the image. These rapidly developing methods for image analysis have resulted in a range of brain-derived measures that can be analyzed statistically, to determine factors that affect brain structure and function, and factors that preserve and promote the integrity of the brain.
Measuring brain integrity
Several different methods are used to image the brain. Each method is unique and can reveal various anatomical, functional, metabolic, or chemical attributes of the brain. Our focus in this review is on recent discoveries made with MRI and diffusion-weighted MRI (dMRI)—by far the most widely used methods for exploring the physical structure of the human brain, and factors that affect it. As noted earlier, standard anatomical MRI—the kind employed most widely in clinical assessments and research studies—is ideal for visualizing patterns and variations in brain structure, especially in the cortical gray matter. Diffusion MRI is more useful for mapping the integrity of white matter fiber tracts and their connectivity. Fractional anisotropy (FA) is one of the most widely accepted measures to represent white matter tissue integrity that is obtainable from DTI. FA, measured on a scale of 0–1, reflects how directionally constrained the diffusion of water is, along axons in a given volume such as a voxel. As a general rule of thumb, higher FA values may imply more coherent or intact axons, or higher degrees of myelination, while lower FA may reflect loss of integrity and white matter injury. Even so, there are known exceptions to this rule in highly convoluted regions of the white matter, because the FA measure appears to be reduced in regions where fibers cross each other, even when the integrity of the fibers is high.
Figure 1 shows various MRI-based features that may be extracted from standard MRI and dMRI techniques. Once these measures are derived, statistical methods from quantitative genetics and epidemiology may be used to map out effects of different dietary, genetic or other factors on these brain measures. Using a database of brain scans, one can study a variety of questions, such as those investigating
A variety of brain measures and features may be extracted from standard MRI and diffusion based MRI. When measured in a population of subjects, the observed variation in these brain traits can be studied in relation to genetic variation, and environmental influences such as education or diet, to identify factors that may promote or harm the brain. Population-based analyses include mapping out the overall degree of genetic influence or heritability for different brain measures, and effects of candidate gene or dietary or environmental factors. Features analyzed may include, but are not limited to: a volumes of brain structures such as the hippocampus; b cortical thickness or folding patterns; c regional brain volume differences with tensor based-morphometry maps; d maps of white matter integrity based on DTI, including measures of fiber coherence, such as fractional anisotropy (FA); e shapes and integrity of tracts of the brain; and f structural brain networks and the strength of cortical connections via white matter fibers, and topological measures based on such networks
-
(a)
the volume of specific structures in the brain, such as the hippocampus [14];
- (b)
-
(c)
regional brain volume differences, assessed using “tensor based-morphometry” maps (TBM; [21]);
-
(d)
maps of white matter integrity such as diffusion tensor imaging (DTI)-based fractional anisotropy (FA) [22];
-
(e)
the trajectories, shapes, and integrity of tracts of the brain [23]; and
-
(f)
the structural human brain network—including the strength of cortical connections via white matter fibers and topological measures based on such networks [24–26].
Psychiatric disorders, such as schizophrenia and bipolar illness, neurological diseases such as Alzheimer’s disease, multiple sclerosis, and even viral infection with the human immunodeficiency virus (HIV) [27] lead to identifiable patterns of anatomical abnormalities and loss of tissue integrity, which are clearly seen in imaging studies. Remarkably, even a person’s diet can have a detectable influence on brain structure and function [28, 29]. Neuroimaging studies confirm that brain integrity is harmed by many drugs of abuse–ecstasy [30], methamphetamine [31, 32]—and excessive alcohol consumption [33]. Perhaps surprisingly, neuroimaging studies also have the power to detect the more subtle effects of specific daily nutrients. If properly designed (see caveats below), they can relate neurological changes and differences in brain integrity to specific dietary factors.
It has been shown, for example, that a group of pre-term infants fed a high-nutrient diet had significantly larger caudate volumes years later as adolescents [34] than those fed a standard diet; males in the high nutrient group of the study also had higher verbal IQ. Before describing more studies of dietary factors and their effects on the brain, we start by noting several major caveats that apply to many epidemiological studies of nutrition and diet, whether or not they use neuroimaging.
Caveats in studying nutrition and the brain
First, in a purely observational study of any population, the intake of any one specific nutrient may be correlated with a host of other dietary factors or habits, making it difficult to isolate the effects of any one factor. However, more perniciously, our intake of any specific food or dietary component—such as sufficient iron or vitamins—may be statistically associated with better (or poorer) diet overall, socio-economic status, or with ethnic or demographic factors. The presence of these confounds makes it crucial, where possible, to control for effects of other possible determinants of brain function that may be driving the associations with a dietary factor.
For example, in studies of fish consumption, both geographical and cultural differences may affect how much fish there is in an individual’s diet (e.g., proximity to the coast). Economic factors are relevant as well. Household income may affect diet, and, depending on the population studied, socio-economic status may correlate with other health-related behaviors, for example with high or low body mass index—which is known to relate to brain structure.
Second, people who make healthy dietary choices tend to make health-promoting choices in other aspects of life, such as increased cardiovascular exercise. They may also avoid excessive alcohol intake or drug abuse, which are detrimental to the brain. Third, there are some associations between educational level and dietary choices, making it difficult to disentangle the primary effects of education on the brain, and effects of education on diet and food choices [35].
As such, it is vital to bear in mind that many studies of dietary factors and the brain are cross-sectional and do not employ a “double-blind” interventional design that introduces a nutritional factor in a randomized way. Such a design would be needed in a clinical trial to make causal statements about specific effects of the nutrient. As such, associations seen cross-sectionally need to be assessed critically. As we will see later, some genetic studies can get around this issue using the concept of “Mendelian randomization” [36]. First, however, we will review a number of studies tracking associations between dietary habits and measurable characteristics of the brain (Table 1). We note that a study not controlling for confounders is not necessarily invalid, as it may report a correlation that truly is present. Care is often needed in critically assessing what may be driving the association, what is the most likely cause, and how likely it is to generalize to new studies or individuals considering dietary changes.
Several studies focus on the consumption of fish, which varies widely across the world. Greater fish consumption is linked to better preservation of neuroanatomical integrity, including reduced white matter abnormalities in an elderly population [37, 38]. Specifically, levels of docosahexaenoic acid (DHA), a form of omega-3 fatty acid, measured from red blood cells, may preserve brain integrity. DHA levels have been associated with greater brain volumes and reduced white matter lesion burden in older, dementia-free adults [39].
Folate and B-vitamins, also sold widely as dietary supplements, show associations with cognitive performance in the elderly, in some but not all studies, as well as to differences in brain structure; vitamin B12 levels have been associated with differences in shapes of brain structures including the hippocampus and caudate [40] as well as gray matter volume in general [41], and white matter lesions [42].
Studies of healthy populations tend to be the best controlled for confounds, but unmodeled factors may drive some of the detected associations. As noted earlier, some cohorts include people who are depressed, or even mildly cognitively impaired, and run the risk of picking up an effect of illness on diet, as being ill may affect overall appetite and dietary choices. Chronic illness can affect body mass index and the ability to exercise or engage in any physical activity, and both affect brain structure and function [35]. People who are becoming ill may also change their diet in an effort to improve it. This can lead to paradoxical effects, whereby beneficial dietary changes may even appear to be associated with poor health or worse outcomes, if people are more inclined to pay attention to their diet when they are ill. Studies of vitamin intake in particular can be confounded by this illness effect.
Small amounts of choline, a nutrient also in the B-vitamin family, are created in our bodies, but the major contributor to our choline levels is our diet. Greater choline intake is associated with better cognitive outcomes in elderly and a reduced overall burden of white matter hyperintensities on MRI [43]. Elevated levels of the amino acid, homocysteine, which depend in turn on our intake of folate and B vitamins, can also affect brain integrity on MRI [44]. People with high homocysteine levels have an increased risk for stroke, Alzheimer’s disease and age-related memory impairment [45] through facilitated build-up of toxic beta–amyloid and tau in the brain [46, 47]. As those with high homocysteine levels have a profile of greater atrophy, early interventions through homocysteine lowering diets and drugs may be considered as means to resist brain atrophy in the elderly. In fact, recently, dietary supplements of B-vitamins, such as folic acid, have been reported to lower the risk for cognitive decline and AD, and they may work by lowering homocysteine levels [48].
By far the most common nutritional disorder in the world is iron deficiency [59], which can adversely affect the developing brain (reviewed in [60]). Elevated iron levels can also promote degenerative disorders in the elderly [61]. The key need for iron homeostasis in the brain and body, and the serious adverse effects when iron levels depart from the normal range, have made it an interesting target of study.
Iron, transferrin, and the human brain
Metal ionsFootnote 1 are increasingly important in helping to understand the development and progression of neurological diseases. As mentioned previously, copper, zinc and iron are all particularly critical for brain development. As with iron, inadequate concentrations of copper can cause anemia and developmental delays, while excess amounts of copper have been associated with neurodegenerative diseases such as Parkinson’s and Alzheimer’s diseases [64]. Additionally, zinc also plays a crucial role in neurodegeneration [65]. However, among the group of metal modifiers, the association of iron to the brain is the one of the most well studied, particularly in the context of additional genetic modifiers. Here, we briefly summarize known associations between brain measures and iron levels in the body (see also reviews by [2, 66–68]).
Iron-deficient diets are associated with poorer cognitive achievement in school-aged children [69]. In regions of the world where iron deficiency anemia is prevalent, such as East Africa, iron supplements can increase motor and language capabilities in children [70]. In children with ADHD, brain iron levels differ in thalamic regions [55]. While several of these studies use T 2* imaging, exact quantification of iron content in the brain tissue is not readily possible in vivo. Indirect measurements and conclusions regarding iron content are obtained from of transverse relaxation times: these measure the decay of the magnetic signal due to perturbations in the magnetic field due to nearby “spins”; this rate of decay is generally different for various types of tissues. The “spin” is a property of the proton, which is used to create the nuclear magnetic resonance signal that we generate and measure with MRI. The decay of the MRI signal can be measured and modeled mathematically using a variety of “relaxation times”, which include T 2*, T 2, and T 2’ [71]. These are all related by the following formula: \((T_{2} *)^{ - 1} = \,\left( {T_{2} } \right)^{ - 1} + \left( {T_{2} '} \right)^{ - 1}\). The relaxation time T 2 is tissue-specific, whereas T 2’ is associated with external field effects and includes all individual contributions from macroscopic and microscopic magnetic field inhomogeneities [72]. Therefore, the T 2* relaxation time depends on the intrinsic T 2 relaxation time as well as all individual macroscopic and microscopic magnetic field inhomogeneities. This measure is therefore affected by disturbances in the global and local magnetic field (i.e., inhomogeneities) as well as water diffusion in tissues, and in the presence of paramagnetic substances; these physical and biological considerations must be taken into account when interpreting results.
Iron deficiency can impair cognitive development, but iron overload also damages the brain. However, this damage is usually evident only later in life. Brain iron regulation is disrupted in several neurodegenerative diseases including Alzheimer’s disease [73], Parkinson’s disease [74], and Huntington’s disease [75], all of which involve abnormally high brain iron concentrations in neuroimaging studies. These high iron concentrations may cause neuronal death [76, 77].
Iron transport into the brain must be carefully regulated, as insufficient or excess iron can have devastating neurocognitive effects. Iron is transported into the brain, and throughout the rest of the body, by the iron-binding protein, transferrin, which regulates iron transport along with specialized transferrin receptors [78].
Transferrin levels can increase in iron-deficient states: when iron levels are low, the liver compensates by producing more transferrin [79], and less transferrin in cases of iron overload [80]. This iron level-dependent fluctuation in transferrin levels means that transferrin itself can serve as a good proxy measure of iron availability to the body. In fact, serum levels of iron fluctuate greatly [81] and depend on dietary factors such as vitamin C intake [82] and the time of blood collection [83]. The gold standard for measuring iron levels in the body is an invasive bone marrow or liver test, but these are not practical to use on volunteers in research studies. On the other hand, transferrin levels are easily measured with a blood test. This gives a reliable and reproducible index of the long-term availability of iron to the brain [84, 85].
Most of the brain’s iron is found in microglia and oligodendrocytes, where it supports myelination [67] and where iron homeostasis is maintained in the brain. Recently, we found that in healthy young adults, fractional anisotropy (FA), the most common measure of white matter integrity computed from DTI scans of the brain, is directly related to serum transferrin levels taken during adolescence [18]. Lower transferrin levels (indicating adequate to higher iron levels) reflected higher FA, or greater brain integrity (Fig. 2). This strong correlation suggests the importance of iron levels in the developing brain, and the ability for non-invasive brain imagingFootnote 2 to trace these effects even in healthy young people.
Using data presented in Jahanshad et al. [18], here we show the association of serum transferrin with a variations in local brain volume, including both positive associations (blue), and negative associations (red/yellow); and b variations in regional measures of fiber integrity as measured through diffusion based fractional anisotropy (FA). All FA associations were negative, such that as serum transferrin increased, regional integrity was reduced
Neuroimaging genetics and iron-related genes
Iron enters the body mainly through the foods we consume. Even so, genetic factors affect the way our bodies absorb and process many nutrients, including iron, through various regulatory proteins involved in its metabolism.
Several genes have been linked to iron homeostasis, including: ACO1, CALR, CD163, CP, CUBN, CYBRD1, DHCR7, EXOC6, FLVCR, FTH1, FTL, FXN, GAST, GSTP1, HAMP, HCP1, HEPH, HEPHL1, HFE, HFE2, HMOX1, HMOX2, HP, HPX, IREB2, PGRMC1, PGRMC2, SLC11A2, SLC25A37, SLC40A1, STEAP3, TF, TFR2, TFRC, TNF [87]. Some of these show associations in multiple ethnic groups [88]. However, not all have been shown to have neurological implications.
Table 2 summarizes some of the genes involved in iron homeostasis that have so far been studied with respect to human brain structure, cognition, or neurological diseases.
The genes presented in the table include HFE, HFE2, HAMP, TFR2, ACO1, FTL, NEO1, TFRC, FPN1, HEPH and TF. Approximately 80 % of hereditary hemochromatosis (HH) is explained by mutations in the HFE gene (or Type 1 HH), but other genes including HFE2, HAMP, TFR2 and FPN-related genes are all involved in non-HFE HH through their direct or indirect regulation of hepcidin, an iron-regulating hormone in the liver [115, 116]. HFE2 is associated with Type 2A juvenile hemochromatosis (JH)-a rare autosomal recessive form of hemochromatosis that causes severe organ damage and premature death before age 40. The non-HFE HH gene, HAMP, is also known to be associated with Type 2B JH; TFR2, expressed almost entirely in the liver, also is involved with hepcidin synthesis and can lead to a form of iron overload that is similar to HFE HH, also without HFE involvement; this is known as Type 3 HH [115]. FPN1, or SLC40A1, codes for the ferroportin protein that transports iron from the inside to the outside of a cell. It is essential in development, and is associated with Type 4 HH, or ferroportin disease, an autosomal dominant disease [115, 116].
ACO1, also known as iron regulatory element binding protein 1, binds to ferritin and transferrin mRNA; polymorphisms in the gene have also been associated with age-related macular degeneration [117].
Variants in the FTL, NEO1, TFRC, and HEPH genes alone are not causes for HH, but these genes are essential in regulating iron homeostasis and transport. FTL is the gene encoding the ferritin light polypeptide, one of the two subunits of the ferritin molecule; it is another gene important for iron homeostasis. Mutations in the gene were first used to describe neuroferritinopathy, evident from iron deposition in the basal ganglia [107]. NEO1, is closely related to a tumor suppressor gene DCC and binds HFE2, and through their interaction, regulates iron homeostasis in hepatocytes and possible skeletal tissue [118, 119]. TFRC encodes the transferrin receptor protein 1 (TfR1)—the protein required for the uptake of transferrin-bound iron in human cells [120]. HEPH or hephaestin, is necessary for transporting iron out of the small intestine’s enterocytes and into the circulation [121].
Some of these studies found no association between the specific candidate genes and the disease of interest, leading to inconsistent evidence, especially for the transferrin gene, TF. Commonly-carried variants in TF (in addition to variants in HFE) accounted for ~40 % of the variance in serum transferrin levels in two normal, healthy populations of approximately 400 individuals [122]. Variants in the TF gene were associated with schizophrenia in a Chinese population of approximately 300 cases and 300 controls [113] but not in a Japanese population with approximately the same number of individuals [114]. Also, comparing about 200 cases to 200 controls, [100] found TF polymorphism G258S was associated with PD, but this effect was not replicated in a Spanish population with roughly the same number of patients [112].
Additionally, the HFE or hemochromatosis gene is associated with iron metabolism disorders. The rarer C282Y mutation in the gene can cause hereditary hemochromatosis (a genetic disorder of iron overload), while the more common H63D variant has been the repeated focus of attention for degenerative brain diseases [123]. Copy number variants in the gene may also be over-represented in people with autism spectrum disorder [102]. Variants in iron-metabolism genes clearly play a role in brain development and degeneration, and this has led to efforts to find molecular pathways by which these genes affect the brain. Brain imaging can facilitate the search for these molecular pathways—by measuring the degree to which these genetic risk factors are associated with brain differences.
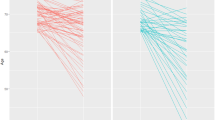
For example, we recently used a study design involving twins—called a cross-twin cross-trait study—to show that commonly carried genetic variations contribute to the normal variation in both transferrin levels and brain integrity as measured with DTI scans In other words, there is pleiotropy—the same genes are implicated in both measures. When individually evaluating all common genetic variants in TF and HFE, variants that together explain ~40 % of the variation in transferrin levels [122], we found that healthy carriers of the H63D polymorphism in the HFE gene have characteristic differences in brain structure, Fig. 3.
Using data presented in Jahanshad et al. [18], we show the association of the H63D polymorphism in the HFE gene with variations in localized fiber integrity, as measured through diffusion-based fractional anisotropy. All FA associations were positive, so the minor allele at H63D showed increases in FA in a dose-dependent manner
This HFE variant is commonly associated with neurodegeneration later in life, but our study of younger people showed higher integrity in healthy carriers of the HFE variant, possibly due to better myelination, approximately 50 years before the average onset age of dementia. However, when examining the same variant in elderly men, Bartzokis et al. [105] found that the presence of the H63D HFE variant (and/or C2 allele of the transferrin gene) was associated with increased basal ganglia iron concentrations compared to noncarriers, suggesting iron overload in the brains of these carriers—a potential risk for degeneration and dementia. Many studies have addressed the association of the H63D variant and the HFE gene on Alzheimer’s disease, with mixed findings [123] regarding the direction of association, the interaction of genes, and the sex most affected. Brain imaging in larger cohorts may help to disentangle the effects of HFE and other iron-related genes on brain structure throughout life.
Conclusion
Neuroimaging genetics now offers great power to understand genetic modulators of dietary influences and their metabolic pathways. By screening brain databases, we may be able to discover nutrients and related genes impacting the brain, deepening our understanding of how certain genes contribute to cognitive outcomes. Additionally, monitoring effects of these mutations with respect to diet as we age may reveal critical periods in the human lifespan when risk for disease is greatest, or when dietary effects are most pronounced. In people at high risk for iron overload disorders, for example, simple preventative nutritional monitoring can help delay, slow down, or even prevent cases of disease.
As we have noted, there are limitations in studies that use brain imaging to map nutritional or genetic effects on the brain. Despite the promise and growth of the field, replication in large samples is needed to boost confidence in the findings. As most single-site imaging studies are small and relatively underpowered to pick up dietary effects or genetic associations, large consortium efforts (e.g., ENIGMA; http://enigma.loni.ucla.edu) are underway to improve power in discovery and replication of subtle effects on the brain. Any actionable factor discovered to resist brain aging or promote brain integrity could impact the lives of hundreds of millions of people worldwide. Many large epidemiological studies now include brain imaging as part of our armory of tools to facilitate this quest.
Notes
At the physiological concentrations present in the brain, copper levels do not appear to influence the signal on standard brain MRI. However, paramagnetic materials such as copper (II) can be and are often used as contrast agents in MRI scans. A newer MRI imaging method, susceptibility-weighted imaging, takes advantage of the distortions in the magnetic field from susceptibility differences in tissue types due to paramagnetic or diamagnetic compounds, to map out cerebrovascular changes in the brain. While this is out of the scope of our current review, we point the reader to the following reference for more information [62, 63].
Non-invasive imaging procedures using MRI have also been developed to map iron concentrations in the brain. T 2* contrast imaging [86] may be used to measure the degree of iron in living tissue. T 2*, and its inverse, R 2, are decay constants that measure of how fast the MRI signal decays, due to magnetic resonance relaxation phenomena. The physics of this process is quite well understood, and iron disrupts the process. As a result, the R 2 relaxation parameter is increased by elevated iron levels in brain tissue. As such, R 2 offers a relative, but not absolute, measure of brain iron levels. Another form of MRI imaging, known as field-dependent R 2 increase (FDRI), uses field-dependent properties of tissues to quantify tissue iron [73]. In vivo evaluation of brain iron in Alzheimer's disease and normal subjects is also possible using MRI. This method may involve scanning a person on two different MRI scanners with different field strengths.
Abbreviations
- MRI:
-
Magnetic resonance imaging
- dMRI:
-
Diffusion magnetic resonance imaging
- DTI:
-
Diffusion tensor imaging
- SNP:
-
Single-nucleotide polymorphism
- FA:
-
Fractional anisotropy
- TBM:
-
Tensor-based morphometry
- DHA:
-
Docosahexaenoic acid
- EPA:
-
Eicosapentaenoic acid
- AD:
-
Alzheimer’s disease
- PD:
-
Parkinson’s disease
- MS:
-
Multiple sclerosis
- RLS:
-
Restless leg syndrome
- ALS:
-
Amyotrophic lateral sclerosis
- SZ:
-
Schizophrenia
- ADHD:
-
Attention deficit hyperactivity disorder
References
Georgieff MK (2007) Nutrition and the developing brain: nutrient priorities and measurement. Am J Clin Nutr 85:614S–620S
Beard JL, Connor JR, Jones BC (1993) Iron in the brain. Nutr Rev 51:157–170
Thompson PM, Jahanshad N (2012) Ironing out neurodegeneration: is iron intake important during the teenage years? Expert Rev Neurother 12:629–631
Thompson PM, Martin NG, Wright MJ (2010) Imaging genomics. Curr Opin Neurol 23:368–373
Braskie MN, Ringman JM, Thompson PM (2011) Neuroimaging measures as endophenotypes in Alzheimer’s disease. Int J Alzheimers Dis 2011:490140
Honea RA, Vidoni E, Harsha A, Burns JM (2009) Impact of APOE on the healthy aging brain: a voxel-based MRI and DTI study. J Alzheimers Dis 18:553–564
Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW et al (2011) Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci 31:6764–6770
Rajagopalan P, Jahanshad N, Stein JL, Kohannim O, Hibar DP, Hua X, Toga AW, J CR Jr, Saykin AJ, Green RC et al (2012) Commonly carried C677T risk variant in the folate pathway candidate gene, MTHFR, promotes brain deficits in the cognitively impaired elderly. NeuroImage-Clin 1:179–187
Sprooten E, Sussmann JE, Moorhead TW, Whalley HC, Ffrench-Constant C, Blumberg HP, Bastin ME, Hall J, Lawrie SM, McIntosh AM (2011) Association of white matter integrity with genetic variation in an exonic DISC1 SNP. Mol Psychiatry 16(685):688–689
McIntosh AM, Moorhead TW, Job D, Lymer GK, Munoz Maniega S, McKirdy J, Sussmann JE, Baig BJ, Bastin ME, Porteous D et al (2008) The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry 13:1054–1059
Braskie MN, Jahanshad N, Stein JL, Barysheva M, Johnson K, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM et al (2012) Relationship of a variant in the NTRK1 gene to white matter microstructure in young adults. J Neurosci 32:5964–5972
Dennis EL, Jahanshad N, Rudie JD, Brown JA, Johnson K, McMahon KL, de Zubicaray GI, Montgomery G, Martin NG, Wright MJ et al (2011) Altered structural brain connectivity in healthy carriers of the autism risk gene, CNTNAP2. Brain Connect 1:447–459
Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, Thompson PM, Geschwind DH, Bookheimer SY, Levitt P et al (2012) Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron 75:904–915
Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O et al (2012) Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44:552–561
Taal HR, St Pourcain B, Thiering E, Das S, Mook-Kanamori DO, Warrington NM, Kaakinen M, Kreiner-Moller E, Bradfield JP, Freathy RM et al (2012) Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet 44:532–538
Ikram MA, Fornage M, Smith AV, Seshadri S, Schmidt R, Debette S, Vrooman HA, Sigurdsson S, Ropele S, Taal HR et al (2012) Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet 44:539–544
Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, Debette S, Shulman JM, Schmidt H, Srikanth V et al (2012) Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet 44:545–551
Jahanshad N, Kohannim O, Hibar DP, Stein JL, McMahon KL, de Zubicaray GI, Medland SE, Montgomery GW, Whitfield JB, Martin NG et al (2012) Brain structure in healthy adults is related to serum transferrin and the H63D polymorphism in the HFE gene. Proc Natl Acad Sci USA 109:E851–E859
Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J et al (2004) Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. Neuroimage 23(Suppl 1):S2–S18
Luders E, Narr KL, Thompson PM, Rex DE, Jancke L, Steinmetz H, Toga AW (2004) Gender differences in cortical complexity. Nat Neurosci 7:799–800
Hua X, Hibar DP, Ching CR, Boyle CP, Rajagopalan P, Gutman BA, Leow AD, Toga AW, Jack CR Jr, Harvey D et al (2013) Unbiased tensor-based morphometry: improved robustness and sample size estimates for Alzheimer’s disease clinical trials. Neuroimage 66C:648–661
Jahanshad N, Kochunov P, Sprooten E, Mandl RC, Nichols TE, Almasy L, Blangero J, Brouwer RM, Curran JE, de Zubicaray GI et al. (2013) Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA–DTI working group. Neuroimage, In Press
Prasad G, Joshi SH, Jahanshad N, Villalon J, Aganj I, Lenglet C, Sapiro G, McMahon KL, de Zubicaray GI, Martin NG (2011) White matter tract analysis in 454 adults using maximum density paths, Proc. MICCAI 2011 Workshop on computational diffusion MRI, September 18–22, 2011
Jahanshad N, Valcour VG, Nir TM, Kohannim O, Busovaca E, Nicolas K, Thompson PM (2012) Disrupted brain networks in the aging HIV + population. Brain Connect 2:335–344
Engel J, Thompson P, Stern J, Staba R, Bragin A, Mody I (2013) Connectomics and epilepsy. Curr Opin Neurol 26:186–194
Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, Jack CR, Saykin AJ, Green RC, Weiner MW et al (2013) Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc Natl Acad Sci USA 110:4768–4773
Nir T, Jahanshad N, Busovaca E, Wendelken L, Nicolas K, Thompson P, Valcour V (2012) Mapping white matter integrity in elderly people with HIV. Human Brain Mapp. In Press
Bourre JM (2006) Effects of nutrients (in food) on the structure and function of the nervous system: update on dietary requirements for brain. Part 2: macronutrients. J Nutr Health Aging 10:386–399
Bourre JM (2006) Effects of nutrients (in food) on the structure and function of the nervous system: update on dietary requirements for brain. Part 1: micronutrients. J Nutr Health Aging 10:377–385
den Hollander B, Schouw M, Groot P, Huisman H, Caan M, Barkhof F, Reneman L (2012) Preliminary evidence of hippocampal damage in chronic users of ecstasy. J Neurol Neurosurg Psychiatry 83:83–85
Thompson PM, Hayashi KM, Simon SL, Geaga JA, Hong MS, Sui Y, Lee JY, Toga AW, Ling W, London ED (2004) Structural abnormalities in the brains of human subjects who use methamphetamine. J Neurosci 24:6028–6036
Sowell ER, Leow AD, Bookheimer SY, Smith LM, O’Connor MJ, Kan E, Rosso C, Houston S, Dinov ID, Thompson PM (2010) Differentiating prenatal exposure to methamphetamine and alcohol versus alcohol and not methamphetamine using tensor-based brain morphometry and discriminant analysis. J Neurosci 30:3876–3885
Pfefferbaum A, Adalsteinsson E, Sullivan EV (2006) Dysmorphology and microstructural degradation of the corpus callosum: interaction of age and alcoholism. Neurobio Aging 27:994–1009
Isaacs EB, Gadian DG, Sabatini S, Chong WK, Quinn BT, Fischl BR, Lucas A (2008) The effect of early human diet on caudate volumes and IQ. Pediatr Res 63:308–314
Ho AJ, Raji CA, Becker JT, Lopez OL, Kuller LH, Hua X, Dinov ID, Stein JL, Rosano C, Toga AW et al (2011) The effects of physical activity, education, and body mass index on the aging brain. Hum Brain Mapp 32:1371–1382
Davey Smith G, Ebrahim S (2003) Mendelian randomization: can genetic epidemiology contribute to understanding environmental determinants of disease? Intern J Epidemiol 32:1–22
Virtanen JK, Siscovick DS, Longstreth WT Jr, Kuller LH, Mozaffarian D (2008) Fish consumption and risk of subclinical brain abnormalities on MRI in older adults. Neurology 71:439–446
Boyle C, Raji CA, Teverovskiy L, Rajagopalan P, Madsen SK, Kuller L, Carmichael OT, Becker JT, Lopez OL, Thompson PM (2012) In organization for human brain mapping (OHBM). Beijing, China
Tan ZS, Harris WS, Beiser AS, Au R, Himali JJ, Debette S, Pikula A, Decarli C, Wolf PA, Vasan RS et al (2012) Red blood cell omega-3 fatty acid levels and markers of accelerated brain aging. Neurology 78:658–664
Ching C, Rajagopalan P, Toga AW, J CR Jr, Weiner MW, Thompson PM (2012) In Proc. society for neuroscience. New Orleans, LA
Erickson KI, Suever BL, Prakash RS, Colcombe SJ, McAuley E, Kramer AF (2008) Greater intake of vitamins B6 and B12 spares gray matter in healthy elderly: a voxel-based morphometry study. Brain Res 1199:20–26
de Lau LM, Smith AD, Refsum H, Johnston C, Breteler MM (2009) Plasma vitamin B12 status and cerebral white-matter lesions. J Neurol Neurosurg Psychiatry 80:149–157
Poly C, Massaro JM, Seshadri S, Wolf PA, Cho E, Krall E, Jacques PF, Au R (2011) The relation of dietary choline to cognitive performance and white-matter hyperintensity in the Framingham offspring cohort. Am J of Clin Nutr 94:1584–1591
Rajagopalan P, Hua X, Toga AW, Jack CR Jr, Weiner MW, Thompson PM (2011) Homocysteine effects on brain volumes mapped in 732 elderly individuals. NeuroReport 22:391–395
Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’Agostino RB, Wilson PW, Wolf PA (2002) Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. New Engl J Med 346:476–483
Mattson MP, Shea TB (2003) Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends Neurosci 26:137–146
Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM (1998) Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch Neurol 55:1449–1455
Smith AD, Smith SM, de Jager CA, Whitbread P, Johnston C, Agacinski G, Oulhaj A, Bradley KM, Jacoby R, Refsum H (2010) Homocysteine-lowering by B vitamins slows the rate of accelerated brain atrophy in mild cognitive impairment: a randomized controlled trial. PLoS One 5:e12244
Conklin SM, Gianaros PJ, Brown SM, Yao JK, Hariri AR, Manuck SB, Muldoon MF (2007) Long-chain omega-3 fatty acid intake is associated positively with corticolimbic gray matter volume in healthy adults. Neurosci Letters 421:209–212
Samieri C, Maillard P, Crivello F, Proust-Lima C, Peuchant E, Helmer C, Amieva H, Allard M, Dartigues JF, Cunnane SC et al (2012) Plasma long-chain omega-3 fatty acids and atrophy of the medial temporal lobe. Neurology 79:642–650
Payne ME, Anderson JJ, Steffens DC (2008) Calcium and vitamin D intakes may be positively associated with brain lesions in depressed and nondepressed elders. Nutr Res 28:285–292
Weinstock-Guttman B, Zivadinov R, Qu J, Cookfair D, Duan X, Bang E, Bergsland N, Hussein S, Cherneva M, Willis L et al (2011) Vitamin D metabolites are associated with clinical and MRI outcomes in multiple sclerosis patients. J Neurol Neurosurg Psychiatry 82:189–195
Buell JS, Dawson-Hughes B, Scott TM, Weiner DE, Dallal GE, Qui WQ, Bergethon P, Rosenberg IH, Folstein MF, Patz S et al (2010) 25-Hydroxyvitamin D, dementia, and cerebrovascular pathology in elders receiving home services. Neurology 74:18–26
Peran P, Cherubini A, Luccichenti G, Hagberg G, Demonet JF, Rascol O, Celsis P, Caltagirone C, Spalletta G, Sabatini U (2009) Volume and iron content in basal ganglia and thalamus. Hum Brain Mapp 30:2667–2675
Cortese S, Azoulay R, Castellanos FX, Chalard F, Lecendreux M, Chechin D, Delorme R, Sebag G, Sbarbati A, Mouren MC et al (2012) Brain iron levels in attention-deficit/hyperactivity disorder: a pilot MRI study. World J Biol Psychiatry 13:223–231
Bartzokis G, Sultzer D, Cummings J, Holt LE, Hance DB, Henderson VW, Mintz J (2000) In vivo evaluation of brain iron in Alzheimer disease using magnetic resonance imaging. Arch Gen Psychiatry 57:47–53
Wang Y, Butros SR, Shuai X, Dai Y, Chen C, Liu M, Haacke EM, Hu J, Xu H (2012) Different iron-deposition patterns of multiple system atrophy with predominant parkinsonism and idiopathetic Parkinson diseases demonstrated by phase-corrected susceptibility-weighted imaging. Am J Neurorad 33:266–273
House MJ, St Pierre TG, Milward EA, Bruce DG, Olynyk JK (2010) Relationship between brain R(2) and liver and serum iron concentrations in elderly men. Mag Reson Med 63:275–281
(2002) Iron deficiency—United States, 1999–2000. MMWR. Morbidity and mortality weekly report, 51, 897–899
Lozoff B, Beard J, Connor J, Barbara F, Georgieff M, Schallert T (2006) Long-lasting neural and behavioral effects of iron deficiency in infancy. Nutrition Rev 64:S34–s43 (Discussion S72-91)
Zecca L, Youdim MB, Riederer P, Connor JR, Crichton RR (2004) Iron, brain ageing and neurodegenerative disorders. Nat Rev Neurosci 5:863–873
Haacke EM, Reichenbach JR (2011) Susceptibility weighted imaging in MRI : basic concepts and clinical applications. Wiley-Blackwell, Hoboken
Fischer A, Hall L (1994) Roles for paramagnetic substances in MRI: contrast agents, molecular amplifiers, and indicators for redox and pH mapping. Mag Res Mater Physics Biol Med 2:203–210
Desai V, Kaler SG (2008) Role of copper in human neurological disorders. Am J Clin Nutrition 88:855S–858S
Frederickson CJ, Koh JY, Bush AI (2005) The neurobiology of zinc in health and disease. Nat Rev Neurosci 6:449–462
Moos T, Rosengren Nielsen T, Skjorringe T, Morgan EH (2007) Iron trafficking inside the brain. J Neurochem 103:1730–1740
Todorich B, Pasquini JM, Garcia CI, Paez PM, Connor JR (2009) Oligodendrocytes and myelination: the role of iron. Glia 57:467–478
Zecca L, Youdim MB, Riederer P, Connor JR, Crichton RR (2004) Iron, brain ageing and neurodegenerative disorders. Nat Rev Neurosci 5:863–873
Halterman JS, Kaczorowski JM, Aligne CA, Auinger P, Szilagyi PG (2001) Iron deficiency and cognitive achievement among school-aged children and adolescents in the United States. Pediatrics 107:1381–1386
Stoltzfus RJ, Kvalsvig JD, Chwaya HM, Montresor A, Albonico M, Tielsch JM, Savioli L, Pollitt E (2001) Effects of iron supplementation and anthelmintic treatment on motor and language development of preschool children in Zanzibar: double blind, placebo controlled study. BMJ 323:1389–1393
Solomon I (1955) Relaxation processes in a system of two spins. Phys Rev 99:559–565
Prince JL, Links JM (2006) Medical imaging signals and systems. Pearson Prentice Hall, Upper Saddle River
Bartzokis G, Sultzer D, Mintz J, Holt LE, Marx P, Phelan CK, Marder SR (1994) In vivo evaluation of brain iron in Alzheimer’s disease and normal subjects using MRI. Biol Psychiatry 35:480–487
Bartzokis G, Cummings JL, Markham CH, Marmarelis PZ, Treciokas LJ, Tishler TA, Marder SR, Mintz J (1999) MRI evaluation of brain iron in earlier- and later-onset Parkinson’s disease and normal subjects. Magn Reson Imag 17:213–222
Jurgens, C.K., Jasinschi, R., Ekin, A., Witjes-Ane, M.N., Middelkoop, H., van der Grond, J. and Roos, R. (2010) MRI T2 Hypointensities in basal ganglia of premanifest Huntington’s disease. PLoS Curr. 2
Ke Y, Ming Qian Z (2003) Iron misregulation in the brain: a primary cause of neurodegenerative disorders. Lancet Neurol 2:246–253
Benarroch EE (2009) Brain iron homeostasis and neurodegenerative disease. Neurology 72:1436–1440
Moos T, Morgan EH (2000) Transferrin and transferrin receptor function in brain barrier systems. Cell Mol Neurobiol 20:77–95
Das Gupta A, Abbi A (2003) High serum transferrin receptor level in anemia of chronic disorders indicates coexistent iron deficiency. Am J Hematol 72:158–161
Khumalo H, Gomo ZA, Moyo VM, Gordeuk VR, Saungweme T, Rouault TA, Gangaidzo IT (1998) Serum transferrin receptors are decreased in the presence of iron overload. Clin Chem 44:40–44
Suominen P, Virtanen A, Lehtonen-Veromaa M, Heinonen OJ, Salmi TT, Alanen M, Mottonen T, Rajamaki A, Irjala K (2001) Regression-based reference limits for serum transferrin receptor in children 6 months to 16 years of age. Clin Chem 47:935–937
Lynch SR, Cook JD (1980) Interaction of vitamin C and iron. Ann NY Acad Sci 355:32–44
Whitfield JB, Cullen LM, Jazwinska EC, Powell LW, Heath AC, Zhu G, Duffy DL, Martin NG (2000) Effects of HFE C282Y and H63D polymorphisms and polygenic background on iron stores in a large community sample of twins. Am J Hum Genet 66:1246–1258
Baynes RD (1996) Assessment of iron status. Clin Biochem 29:209–215
Ahluwalia N (1998) Diagnostic utility of serum transferrin receptors measurement in assessing iron status. Nutr Rev 56:133–141
Drayer B, Burger P, Darwin R, Riederer S, Herfkens R, Johnson GA (1986) MRI of brain iron. Am J Roentgenol 147:103–110
Constantine CC, Gurrin LC, McLaren CE, Bahlo M, Anderson GJ, Vulpe CD, Forrest SM, Allen KJ, Gertig DM (2008) SNP selection for genes of iron metabolism in a study of genetic modifiers of hemochromatosis. BMC Med Genet 9:18
McLaren CE, McLachlan S, Garner CP, Vulpe CD, Gordeuk VR, Eckfeldt JH, Adams PC, Acton RT, Murray JA, Leiendecker-Foster C et al (2012) Associations between single-nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS One 7:e38339
Hanninen MM, Haapasalo J, Haapasalo H, Fleming RE, Britton RS, Bacon BR, Parkkila S (2009) Expression of iron-related genes in human brain and brain tumors. BMC Neurosci 10:36
Spiegel R, Pines O, Ta-Shma A, Burak E, Shaag A, Halvardson J, Edvardson S, Mahajna M, Zenvirt S, Saada A et al (2012) Infantile cerebellar-retinal degeneration associated with a mutation in mitochondrial aconitase, ACO2. Am J Hum Genet 90:518–523
Lin M, Zhao L, Fan J, Lian XG, Ye JX, Wu L, Lin H (2012) Association between HFE polymorphisms and susceptibility to Alzheimer’s disease: a meta-analysis of 22 studies including 4,365 cases and 8,652 controls. Mol Biol Rep 39:3089–3095
Blazquez L, De Juan D, Ruiz-Martinez J, Emparanza JI, Saenz A, Otaegui D, Sistiaga A, Martinez-Lage P, Lamet I, Samaranch L et al (2007) Genes related to iron metabolism and susceptibility to Alzheimer’s disease in Basque population. Neurobio Aging 28:1941–1943
Guerreiro RJ, Bras JM, Santana I, Januario C, Santiago B, Morgadinho AS, Ribeiro MH, Hardy J, Singleton A, Oliveira C (2006) Association of HFE common mutations with Parkinson’s disease, Alzheimer’s disease and mild cognitive impairment in a Portuguese cohort. BMC Neurol 6:24
Candore G, Licastro F, Chiappelli M, Franceschi C, Lio D, Rita Balistreri C, Piazza G, Colonna-Romano G, Grimaldi LM, Caruso C (2003) Association between the HFE mutations and unsuccessful ageing: a study in Alzheimer’s disease patients from northern Italy. Mech Ageing Dev 124:525–528
Sutedja NA, Sinke RJ, Van Vught PW, Van der Linden MW, Wokke JH, Van Duijn CM, Njajou OT, Van der Schouw YT, Veldink JH, Van den Berg LH (2007) The association between H63D mutations in HFE and amyotrophic lateral sclerosis in a Dutch population. Arch Neurol 64:63–67
He X, Lu X, Hu J, Xi J, Zhou D, Shang H, Liu L, Zhou H, Yan B, Yu L et al (2011) H63D polymorphism in the hemochromatosis gene is associated with sporadic amyotrophic lateral sclerosis in China. Euro J Neurol 18:359–361
Praline J, Blasco H, Vourc’h P, Rat V, Gendrot C, Camu W, Andres CR (2012) Study of the HFE gene common polymorphisms in French patients with sporadic amyotrophic lateral sclerosis. J Neurol Sci 317:58–61
van Rheenen, W., Diekstra, F.P., van Doormaal, P.T., Seelen, M., Kenna, K., McLaughlin, R., Shatunov, A., Czell, D., van Es, M.A., van Vught, P.W. et al. (2012) H63D polymorphism in HFE is not associated with amyotrophic lateral sclerosis. Neurobio Aging. In Press
Gemmati D, Zeri G, Orioli E, De Gaetano FE, Salvi F, Bartolomei I, D’Alfonso S, Dall’osso C, Leone MA, Singh AV et al (2012) Polymorphisms in the genes coding for iron binding and transporting proteins are associated with disability, severity, and early progression in multiple sclerosis. BMC Med Genet 13:70
Borie C, Gasparini F, Verpillat P, Bonnet AM, Agid Y, Hetet G, Brice A, Durr A, Grandchamp B (2002) Association study between iron-related genes polymorphisms and Parkinson’s disease. J Neurol 249:801–804
Oexle K, Schormair B, Ried JS, Czamara D, Heim K, Frauscher B, Hogl B, Trenkwalder C, Martin Fiedler G, Thiery J et al (2012) Dilution of candidates: the case of iron-related genes in restless legs syndrome. Eur J Hum Genet 21:410–414
Rosenfeld JA, Ballif BC, Torchia BS, Sahoo T, Ravnan JB, Schultz R, Lamb A, Bejjani BA, Shaffer LG (2010) Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med 12:694–702
Gebril OH, Meguid NA (2011) HFE gene polymorphisms and the risk for autism in Egyptian children and impact on the effect of oxidative stress. Dis Markers 31:289–294
Bartzokis G, Lu PH, Tishler TA, Peters DG, Kosenko A, Barrall KA, Finn JP, Villablanca P, Laub G, Altshuler LL et al (2010) Prevalent iron metabolism gene variants associated with increased brain ferritin iron in healthy older men. J Alzheimers Dis 20:333–341
Bartzokis G, Lu PH, Tingus K, Peters DG, Amar CP, Tishler TA, Finn JP, Villablanca P, Altshuler LL, Mintz J et al (2011) Gender and iron genes may modify associations between brain iron and memory in healthy aging. Neuropsychopharmacology 36:1375–1384
Kauwe JS, Bertelsen S, Mayo K, Cruchaga C, Abraham R, Hollingworth P, Harold D, Owen MJ, Williams J, Lovestone S et al (2010) Suggestive synergy between genetic variants in TF and HFE as risk factors for Alzheimer’s disease. Am J Med Genet B Neuropsychiatr genet 153B:955–959
Curtis AR, Fey C, Morris CM, Bindoff LA, Ince PG, Chinnery PF, Coulthard A, Jackson MJ, Jackson AP, McHale DP et al (2001) Mutation in the gene encoding ferritin light polypeptide causes dominant adult-onset basal ganglia disease. Nat Genet 28:350–354
Zhang P, Yang Z, Zhang C, Lu Z, Shi X, Zheng W, Wan C, Zhang D, Zheng C, Li S et al (2003) Association study between late-onset Alzheimer’s disease and the transferrin gene polymorphisms in Chinese. Neurosci Lett 349:209–211
Schjeide BM, McQueen MB, Mullin K, DiVito J, Hogan MF, Parkinson M, Hooli B, Lange C, Blacker D, Tanzi RE et al (2009) Assessment of Alzheimer’s disease case-control associations using family-based methods. Neurogenetics 10:19–25
Laumet G, Chouraki V, Grenier-Boley B, Legry V, Heath S, Zelenika D, Fievet N, Hannequin D, Delepine M, Pasquier F et al (2010) Systematic analysis of candidate genes for Alzheimer’s disease in a French, genome-wide association study. J Alzheimers Dis 20:1181–1188
Van Landeghem GF, Sikstrom C, Beckman LE, Adolfsson R, Beckman L (1998) Transferrin C2, metal binding and Alzheimer’s disease. NeuroReport 9:177–179
Ezquerra M, Campdelacreu J, Munoz E, Tolosa E (2005) Association study of the G258S transferrin gene polymorphism and Parkinson’s disease in the Spanish population. J Neurology 252:1269–1270
Qu M, Yue W, Tang F, Wang L, Han Y, Zhang D (2008) Polymorphisms of Transferrin gene are associated with schizophrenia in Chinese Han population. J Psychiatric Res 42:877–883
Maeno N, Takahashi N, Saito S, Ji X, Branko A, Ishihara R, Yoshida K, Inada T, Iidaka T, Ozaki N (2007) Association study between the transferrin gene and schizophrenia in the Japanese population. NeuroReport 18:517–520
Santos PC, Dinardo CL, Cancado RD, Schettert IT, Krieger JE, Pereira AC (2012) Non-HFE hemochromatosis. Rev Bras Hematol Hemoter 34:311–316
Pietrangelo A, Caleffi A, Corradini E (2011) Non-HFE hepatic iron overload. Semin Liver Dis 31:302–318
Synowiec E, Pogorzelska M, Blasiak J, Szaflik J, Szaflik JP (2012) Genetic polymorphism of the iron-regulatory protein-1 and -2 genes in age-related macular degeneration. Mol Biol Rep 39:7077–7087
Yang F, West AP Jr, Bjorkman PJ (2011) Crystal structure of a hemojuvelin-binding fragment of neogenin at 1.8A. J Struct Biol 174:239–244
Lin L, Goldberg YP, Ganz T (2005) Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood 106:2884–2889
Herbison CE, Thorstensen K, Chua AC, Graham RM, Leedman P, Olynyk JK, Trinder D (2009) The role of transferrin receptor 1 and 2 in transferrin-bound iron uptake in human hepatoma cells. Am J Physiol Cell Physiol 297:C1567–C1575
Chen H, Su T, Attieh ZK, Fox TC, McKie AT, Anderson GJ, Vulpe CD (2003) Systemic regulation of Hephaestin and Ireg1 revealed in studies of genetic and nutritional iron deficiency. Blood 102:1893–1899
Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, Palotie A, Peltonen L, Martin NG, Montgomery GW, Whitfield JB et al (2009) Variants in TF and HFE explain approximately 40% of genetic variation in serum–transferrin levels. Am J Hum Genet 84:60–65
Nandar W, Connor JR (2011) HFE gene variants affect iron in the brain. J Nutr 141:729S–739S
Acknowledgments
We are very grateful for the support of our colleagues and co-authors at the UCLA School of Medicine and the University of Queensland who helped to design and perform many of the brain imaging studies reviewed here. This work was funded in part by NIH grants R01 NS080655, R01 MH097268, R01 AG040060, R01 EB008432 and P41 EB015922.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Jahanshad, N., Rajagopalan, P. & Thompson, P.M. Neuroimaging, nutrition, and iron-related genes. Cell. Mol. Life Sci. 70, 4449–4461 (2013). https://doi.org/10.1007/s00018-013-1369-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1369-2