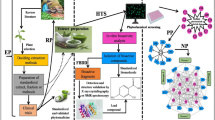
Abstract
Objectives
The aim was to index natural products for less expensive preventive or curative anti-inflammatory therapeutic drugs.
Materials
A set of 441 anti-inflammatory drugs representing the active domain and 2892 natural products representing the inactive domain was used to construct a predictive model for bioactivity-indexing purposes.
Method
The model for indexing the natural products for potential anti-inflammatory activity was constructed using the iterative stochastic elimination algorithm (ISE). ISE is capable of differentiating between active and inactive anti-inflammatory molecules.
Results
By applying the prediction model to a mix set of (active/inactive) substances, we managed to capture 38% of the anti-inflammatory drugs in the top 1% of the screened set of chemicals, yielding enrichment factor of 38. Ten natural products that scored highly as potential anti-inflammatory drug candidates are disclosed. Searching the PubMed revealed that only three molecules (Moupinamide, Capsaicin, and Hypaphorine) out of the ten were tested and reported as anti-inflammatory. The other seven phytochemicals await evaluation for their anti-inflammatory activity in wet lab.
Conclusion
The proposed anti-inflammatory model can be utilized for the virtual screening of large chemical databases and for indexing natural products for potential anti-inflammatory activity.









Similar content being viewed by others
References
Murakami M, Hirano T. The molecular mechanisms of chronic inflammation development. Front Immunol. 2012;3:323. doi:10.3389/fimmu.2012.00323.
Zeinali M, Rezaee SA, Hosseinzadeh H. An overview on immunoregulatory and anti-inflammatory properties of chrysin and flavonoids substances. Biomed Pharmacother. 2017;92:998–1009. doi:10.1016/j.biopha.2017.06.003.
Schwager J, Richard N, Widmer F, Raederstorff D. Resveratrol distinctively modulates the inflammatory profiles of immune and endothelial cells. BMC Complement Altern Med. 2017;17(1):309. doi:10.1186/s12906-017-1823-z.
Mkhize NVP, Qulu L, Mabandla MV. The effect of quercetin on pro- and anti-inflammatory cytokines in a prenatally stressed rat model of febrile seizures. J Exp Neurosci. 2017;11:1179069517704668. doi:10.1177/1179069517704668.
Fenni S, Hammou H, Astier J, Bonnet L, Karkeni E, Couturier C, et al. Lycopene and tomato powder supplementation similarly inhibit high-fat diet induced obesity, inflammatory response, and associated metabolic disorders. Mol Nutr Food Res. 2017;. doi:10.1002/mnfr.201601083.
Dai SX, Li WX, Li GH, Huang JF. Proteome-wide prediction of targets for aspirin: new insight into the molecular mechanism of aspirin. PeerJ. 2016;4:e1791. doi:10.7717/peerj.1791.
Filannino P, Cavoski I, Thlien N, Vincentini O, De Angelis M, Silano M, et al. Lactic acid fermentation of cactus cladodes (Opuntia ficus-indica L.) generates flavonoid derivatives with antioxidant and anti-inflammatory properties. PLoS One. 2016;11(3):e0152575. doi:10.1371/journal.pone.0152575.
Kuo PC, Liao YR, Hung HY, Chuang CW, Hwang TL, Huang SC, et al. Anti-inflammatory and neuroprotective constituents from the peels of Citrus grandis. Molecules. 2017;. doi:10.3390/molecules22060967.
Lee E, Kim SG, Park NY, Park HH, Jeong KT, Choi J, et al. Anti-inflammatory effects of KOTMIN13: a mixed herbal medicine in LPS-stimulated RAW 264.7 cells and mouse edema models. Pharmacogn Mag. 2017;13(50):216–21. doi:10.4103/0973-1296.204548.
Kacergius T, Abu-Lafi S, Kirkliauskiene A, Gabe V, Adawi A, Rayan M, et al. Inhibitory capacity of Rhus coriaria L. extract and its major component methyl gallate on Streptococcus mutants biofilm formation by optical profilometry: potential applications for oral health. Mol Med Rep. 2017;16(1):949–56. doi:10.3892/mmr.2017.6674.
Frank A, Abu-Lafi S, Adawi A, Schwed JS, Stark H, Rayan A. From medicinal plant extracts to defined chemical compounds targeting the histamine H4 receptor: Curcuma longa in the treatment of inflammation. Inflamm Res. 2017;. doi:10.1007/s00011-017-1075-x.
Dutartre P. Inflammasomes and natural ingredients towards new anti-inflammatory agents. Molecules. 2016;. doi:10.3390/molecules21111492.
Paoletta S, Steventon GB, Wildeboer D, Ehrman TM, Hylands PJ, Barlow DJ. Screening of herbal constituents for aromatase inhibitory activity. Bioorg Med Chem. 2008;16(18):8466–70.
Shahaf N, Pappalardo M, Basile L, Guccione S, Rayan A. How to choose the suitable template for homology modelling of GPCRs: 5-HT7 receptor as a test case. Mol Inform. 2016;35(8–9):414–23. doi:10.1002/minf.201501029.
Pappalardo M, Rayan M, Abu-Lafi S, Leonardi ME, Milardi D, Guccione S, et al. Homology-based modeling of rhodopsin-like family members in the inactive state: structural analysis and deduction of tips for modeling and optimization. Mol Inform. 2017;. doi:10.1002/minf.201700014.
Luksch T, Chan NS, Brass S, Sotriffer CA, Klebe G, Diederich WE. Computer-aided design and synthesis of nonpeptidic plasmepsin II and IV inhibitors. ChemMedChem. 2008;3(9):1323–36.
Schuller A, Schneider G. Identification of hits and lead structure candidates with limited resources by adaptive optimization. J Chem Inf Model. 2008;48(7):1473–91.
Zaheer-ul H, Uddin R, Yuan H, Petukhov PA, Choudhary MI, Madura JD. Receptor-based modeling and 3D-QSAR for a quantitative production of the butyrylcholinesterase inhibitors based on genetic algorithm. J Chem Inf Model. 2008;48(5):1092–103.
Hao M, Zhang S, Qiu J. Toward the prediction of FBPase inhibitory activity using chemoinformatic methods. Int J Mol Sci. 2012;13(6):7015–37. doi:10.3390/ijms13067015.
Li H, Yap CW, Ung CY, Xue Y, Li ZR, Han LY, et al. Machine learning approaches for predicting compounds that interact with therapeutic and ADMET related proteins. J Pharm Sci. 2007;96(11):2838–60.
Lusci A, Pollastri G, Baldi P. Deep architectures and deep learning in chemoinformatics: the prediction of aqueous solubility for drug-like molecules. J Chem Inf Model. 2013;53(7):1563–75. doi:10.1021/ci400187y.
Plewczynski D, von Grotthuss M, Spieser SA, Rychlewski L, Wyrwicz LS, Ginalski K, et al. Target specific compound identification using a support vector machine. Comb Chem High Throughput Screen. 2007;10(3):189–96.
Heikamp K, Bajorath J. Comparison of confirmed inactive and randomly selected compounds as negative training examples in support vector machine-based virtual screening. J Chem Inf Model. 2013;53(7):1595–601. doi:10.1021/ci4002712.
Shen M, Beguin C, Golbraikh A, Stables JP, Kohn H, Tropsha A. Application of predictive QSAR models to database mining: identification and experimental validation of novel anticonvulsant compounds. J Med Chem. 2004;47(9):2356–64.
Rayan A. New vistas in GPCR 3D structure prediction. J Mol Model. 2010;16(2):183–91.
Efremov RG, Chugunov AO, Pyrkov TV, Priestle JP, Arseniev AS, Jacoby E. Molecular lipophilicity in protein modeling and drug design. Curr Med Chem. 2007;14(4):393–415.
Deeb O, Goodarzi M. Exploring QSARs for inhibitory activity of non-peptide HIV-1 protease inhibitors by GA-PLS and GA-SVM. Chem Biol Drug Des. 2010;75(5):506–14. doi:10.1111/j.1747-0285.2010.00953.x.
Deeb O, Jawabreh S, Goodarzi M. Exploring QSARs of vascular endothelial growth factor receptor-2 (VEGFR-2) tyrosine kinase inhibitors by MLR, PLS and PC-ANN. Curr Pharm Des. 2013;19(12):2237–44.
Mussa HY, Hawizy L, Nigsch F, Glen RC. Classifying large chemical data sets: using a regularized potential function method. J Chem Inf Model. 2011;51(1):4–14. doi:10.1021/ci100022u.
Glick M, Goldblum A. A novel energy-based stochastic method for positioning polar protons in protein structures from X-rays. Proteins. 2000;38(3):273–87. doi:10.1002/(SICI)1097-0134(20000215)38:3<273:AID-PROT4>3.0.CO;2-I.
Glick M, Rayan A, Goldblum A. A stochastic algorithm for global optimization and for best populations: a test case of side chains in proteins. Proc Natl Acad Sci USA. 2002;99(2):703–8. doi:10.1073/pnas.022418199.
Michaeli A, Rayan A. Modeling ensembles of loop conformations by iterative stochastic elimination. Lett Drug Des Discov. 2016;13(3):1–6.
Rayan A, Senderowitz H, Goldblum A. Exploring the conformational space of cyclic peptides by a stochastic search method. J Mol Gr Model. 2004;22(5):319–33.
Rayan A, Noy E, Chema D, Levitzki A, Goldblum A. Stochastic algorithm for kinase homology model construction. Curr Med Chem. 2004;11(6):675–92.
Rayan A, Marcus D, Goldblum A. Predicting oral druglikeness by iterative stochastic elimination. J Chem Inf Model. 2010;50(3):437–45. doi:10.1021/ci9004354.
Rayan A, Falah M, Raiyn J, Da’adoosh B, Kadan S, Zaid H, et al. Indexing molecules for their hERG liability. Eur J Med Chem. 2013;65C:304–14. doi:10.1016/j.ejmech.2013.04.059.
Pappalardo M, Shachaf N, Basile L, Milardi D, Zeidan M, Raiyn J, et al. Sequential application of ligand and structure based modeling approaches to index chemicals for their hH4R antagonism. PLoS One. 2014;9(10):e109340. doi:10.1371/journal.pone.0109340.
Zaid H, Raiyn J, Osman M, Falah M, Srouji S, Rayan A. In silico modeling techniques for predicting the tertiary structure of human H4 receptor. Front Biosci (Landmark Ed). 2016;21:597–619.
Ren X, Zhang M, Chen L, Zhang W, Huang Y, Luo H, et al. The anti-inflammatory effects of Yunnan Baiyao are involved in regulation of the phospholipase A2/arachidonic acid metabolites pathways in acute inflammation rat model. Mol Med Rep. 2017;. doi:10.3892/mmr.2017.7104.
Zatsepin M, Mattes A, Rupp S, Finkelmeier D, Basu A, Burger-Kentischer A, et al. Computational discovery and experimental confirmation of TLR9 receptor antagonist leads. J Chem Inf Model. 2016;56(9):1835–46. doi:10.1021/acs.jcim.6b00070.
Rayan A. New tips for structure prediction by comparative modeling. Bioinformation. 2009;3(6):263–7.
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26.
Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol. 2004;8(3):255–63. doi:10.1016/j.cbpa.2004.04.003.
Jiang Y, Yu L, Wang MH. N-trans-feruloyltyramine inhibits LPS-induced NO and PGE2 production in RAW 264.7 macrophages: involvement of AP-1 and MAP kinase signalling pathways. Chem Biol Interact. 2015;235:56–62. doi:10.1016/j.cbi.2015.03.029.
Park JB. Isolation and characterization of N-feruloyltyramine as the P-selectin expression suppressor from garlic (Allium sativum). J Agric Food Chem. 2009;57(19):8868–72. doi:10.1021/jf9018382.
Gooding SM, Canter PH, Coelho HF, Boddy K, Ernst E. Systematic review of topical capsaicin in the treatment of pruritus. Int J Dermatol. 2010;49(8):858–65. doi:10.1111/j.1365-4632.2010.04537.x.
Gevorgyan A, Segboer C, Gorissen R, van Drunen CM, Fokkens W. Capsaicin for non-allergic rhinitis. Cochrane Database Syst Rev. 2015;7:CD010591. doi:10.1002/14651858.CD010591.pub2.
Toyoda T, Shi L, Takasu S, Cho YM, Kiriyama Y, Nishikawa A, et al. Anti-inflammatory effects of capsaicin and piperine on Helicobacter pylori-induced chronic gastritis in mongolian gerbils. Helicobacter. 2016;21(2):131–42. doi:10.1111/hel.12243.
Sun H, Cai W, Wang X, Liu Y, Hou B, Zhu X, et al. Vaccaria hypaphorine alleviates lipopolysaccharide-induced inflammation via inactivation of NFkappaB and ERK pathways in Raw 264.7 cells. BMC Complement Altern Med. 2017;17(1):120. doi:10.1186/s12906-017-1635-1.
Lin CF, Hwang TL, Chien CC, Tu HY, Lay HL. A new hydroxychavicol dimer from the roots of Piper betle. Molecules. 2013;18(3):2563–70. doi:10.3390/molecules18032563.
Ganguly S, Mula S, Chattopadhyay S, Chatterjee M. An ethanol extract of Piper betle Linn. mediates its anti-inflammatory activity via down-regulation of nitric oxide. J Pharm Pharmacol. 2007;59(5):711–8. doi:10.1211/jpp.59.5.0012.
Acknowledgements
This work was partially supported by the Al-Qasemi Research Foundation (Grant no. 954000) and the Ministry of Science, Space and Technology, Israel. We declare that the funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: John Di Battista.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aswad, M., Rayan, M., Abu-Lafi, S. et al. Nature is the best source of anti-inflammatory drugs: indexing natural products for their anti-inflammatory bioactivity. Inflamm. Res. 67, 67–75 (2018). https://doi.org/10.1007/s00011-017-1096-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-017-1096-5