Abstract
Official control samples of wheat and rye flour were analyzed for the presence of Shiga-toxin producing Escherichia coli (STEC) in 25 g. The detection procedure was based on enrichment in buffered peptone water and Tryptone Bile X-Glucuronide agar, followed by multiplex real-time PCR with an internal positive control. Positive samples were sub-cultured for strain isolation by a two-step procedure. In the first step, ten colonies were picked from each plate, pooled, and analyzed by real-time PCR. In addition, a bacterial material from the part with heavy growth was analyzed. If the 1st isolation attempt failed, in a second step all colonies from molecular positive plates were picked, sub-cultured, pooled and analyzed. 39% of test samples were positive by real-time PCR. STEC was isolated from 17 test samples corresponding to 19%. Molecular detection is linked to the presence of quantifiable numbers of E. coli, whereas the grain species did not have an influence. There is a non-significant correlation with some mills indicating some technological or hygienic problems in those facilities. Increasing the number of subsamples did improve the detection rate. Our findings show the frequent presence of STEC in flour in Germany.
Similar content being viewed by others
1 Introduction
Shiga-Toxin producing Escherichia coli (STEC) is a major cause of severe human gastroenteritis (Karmali 2004; EFSA Panel on Biological Hazrads (BIOHAZ) 2013; Karch et al. 2015). STEC frequently occurs in food of animal origin, but increasingly its relevance for food of non-animal origin is gaining importance. The STEC outbreak in Germany by contaminated fenugreek sprouts with more than 50 fatal cases has highlighted the problem (Robert Koch Institut 2011). Consequently, official monitoring programs in Germany were initiated for detection STEC in food of non-animal origin (FNAO). The prevalence of STEC in FNAO reported was very low (BVL 2014) or not detectable (BVL 2015, 2016a, b).
Since 2016, reports of the presence of STEC in contaminated flour have been published (CDC 2016; Gieraltowski et al. 2017; Wu et al. 2017). A U.S. company initiated a voluntary recall of wheat flour and products contaminated with STEC O121 and O26 (CDC 2016). Neil et al. (2012) reported STEC in uncooked cookie dough first, but could not trace back this contamination to flour unambiguously. These findings highlight the risk of pathogenic STEC in flour and flour products. Although most wheat and rye flour is being consumed baked, consumption of uncooked flour might occur. New products with uncooked cookie dough, like ice cream, have recently appeared on the market. In households, risky consumption of raw cookie dough or cake batter is quite common. In the US, 53% of young adults ate homemade cookie dough (Byrd-Bredbenner et al. 2008). In addition, flour is often used to dust surfaces for the transfer of bakery products and pizzas (Gieraltowski et al. 2017). Thicker dough could be undercooked at some locations. As wheat and rye flour is a stable food, the very high number of servings may pose a risk of ingesting undercooked flour. (Gilbert et al. 2010; Neil et al. 2012; Gieraltowski et al. 2017; Wu et al. 2017).
Reports of microbial contamination of wheat or rye grains are rare. Microbial contamination of cereal products can occur before or during harvest, or at the flourmill (Gilbert et al. 2010). Berghofer et al. (2003) reported that contamination at the mills is most probably the result of contamination from encrusted grain dust and residues inside the bins. The milling process includes a conditioning step using water to increase the aw from 0.40 to 0.65 (8–12% moisture) to 0.68–0.70 (14–15% moisture). Water is added by means of a fine spray. Conditioned wheat is held in large conditioning bins, usually overnight, but sometimes for up to 24–36 h, depending on wheat type and initial moisture content.
Pathogenic Enterobacteriaceae may occur in products with low water activity (aw) and that exhibits higher heat tolerance (Gilbert et al. 2010; Finn et al. 2013; Beuchat et al. 2013). Thermal inactivation of STEC in low moisture foods requires higher temperatures and is strain dependent (Liu et al. 2014). At temperatures of 70 °C, Escherichia coli O157:H7 that was originally isolated from cookie dough was detectable in flour after 30 min (Greene 2012). Under stress, numerous pathogenic bacteria can enter the viable but non culturable stage (VBNC); bacteria enter a metabolically dormant state and consequently are not culturable using conventional laboratory protocols (Finn et al. 2013; Knödler et al. 2016).
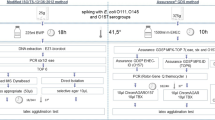
Molecular detection and strain isolation of STEC in food is described in ISO/TS 13136:2012 (ISO 2012). The detection procedure according to this standard comprises enrichment in modified tryptic soy broth (mTSB) or buffered peptone water (BPW), followed by nucleic acid extraction and real-time PCR. Information on real-time PCR systems to be used is given in an informative annex; this opens the possibility to use other systems if considered necessary. Several protocols for detection of stx1-, stx2-, and eae-gene by real-time PCR were published (Reischl et al. 2002; Nielsen and Andersen 2003; Perelle et al. 2004; Pavlovic et al. 2010). The isolation procedure is described in a normative annex B of ISO/TS 13136:2012 and is based on the test of 50 colonies. It became evident that this procedure is not successful when applied to FNAO. Taking this into consideration, modifications of ISO/TS 13136:2012 were introduced into the methods developed for the Official Collection of Methods of Analysis according to § 64 of the German Food and Feed Act (LFGB) (Tzschoppe et al. 2012; Working Group “Molecular Methods—Microbiology” 2012). The non-selective pre-enrichment step in BPW is followed by a selective enrichment step on solid agar. 100 µl pre-enrichment broth is streaked out onto Tryptone Bile X-Glucuronide (TBX) agar in duplicate and incubated at 44 °C. The colonies from TBX agar plates are swept off and subjected to real-time PCR. A combination of non-selective pre-enrichment and selective enrichment allows for the successful detection of sub-lethally injured bacteria (Clark and Ordal 1969; Hoorfar and Baggesen 1998). A selective enrichment at 44 °C suppresses background flora, ensuring reliable real-time PCR results and facilitates isolation procedures (Tzschoppe et al. 2012).
Sequence analysis of STEC isolated from plants did not show any clustering; this indicates that plant specific genotypes do not exist and the presence of STEC in FNAO is caused by contamination (Bauwens et al. 2016). In the framework of official food surveillance programs, flour is being analyzed microbiologically. We detected STEC in 2015 for the 1st time; these findings were verified by follow up studies, and inspections were carried out. This paper describes the molecular detection and microbial isolation of STEC in flour samples.
2 Materials and methods
2.1 Flour samples and microbial analysis
Flour samples were taken as part of official food surveillance programs in Saxony-Anhalt from 2014 to 2017. Local authorities decided on the number of subsamples to be taken based on previous test results and the accessibility of the material. Samples were transported to the laboratory within 48 h at 2 °C. All subsamples were analyzed separately for E. coli and STEC; hence 51 incoming laboratory samples resulted 98 test samples (see Table 1). Using standard protocols, incoming samples were analyzed for total plate count, Salmonella spp., E. coli count, Bacillus cereus, sulfite-reducing clostridia, yeasts and moulds (BVL 1991; ISO 2001, 2003, 2004, 2013). The detection protocol for E. coli was modified by using Brilliance E. coli/Coliform Selective Agar (Oxoid, Wesel, Germany, PO5176A) and incubation at 37 °C. A detailed summary of flour samples and microbial results is given as electronic supplementary material.
2.2 Analysis of STEC
Analysis of STEC is carried out through a combination of the following three modules:
-
1.
microbial enrichment as two step procedure,
-
2.
molecular detection by real-time PCR, and
-
3.
STEC isolation.
2.2.1 Module 1: microbial enrichment
A 25 g portion of flour was added to 225 mL BPW (Oxoid, BO0201S) and incubated for 16 ± 1 h at 37 °C. 100 µL of incubated pre-enrichment broth was streaked onto TBX agar (Oxoid, BO0194SM) in duplicates and incubated at 44 °C for 24 ± 1 h. The next day, 1 mL 0.9% NaCl solution was applied onto the surface of each TBX agar plate and colonies were removed completely by wiping with the pipette tip. The resulting suspension was aspirated with a pipette, and transferred into a reaction tube. 100 µL were removed and diluted by adding 900 µL Aqua bidest, subjected to thermal lysis at 95 °C for 10 min followed by 30 s centrifugation (Tzschoppe et al. 2012; Working Group “Molecular Methods—Microbiology” 2012). The residual bacterial suspension was kept for STEC isolation, if molecular reactions showed the presence of stx genes.
2.2.2 Module 2: multiplex real-time PCR assays
Multiplex real-time PCR was a combination of published systems for the detection of stx1, stx2, eae simultaneously together with an internal amplifications control based on pUC18/pUC19. Real-time PCR assays were carried out in 25 µL reaction volumes containing 2.5 µL of template lysate, 300 nM each primer stx-F, stx-R, eae-F, ear-R, 125 nM each probe stx1-probe; stx2-probe, eae-probe, 250 nM each primer pUC 18-F and pUC 18-R, 100 nM probe Tm-pUC18, 1 µL diluted pUC19 DNA equivalent to approximately 10 copies, and 12.5 µL Roche LightCycler® 480 Probes Master (Roche Diagnostics, Mannheim, Germany). All primers and probes were synthesized by TIB MOLBIOL (Berlin, Germany) and are listed in Table 2. Thermal cycling was carried out on a LightCycler® LC480 (Roche Diagnostics), and consisted of an initial step at 95 °C for 10 min followed by 45 cycles of 95 °C for 20 s and 60 °C for 1 min. Fluorescence acquisition took place at the end of each 60 °C cycle. Reactions were analyzed at wavelengths of 500 nm (stx1), 533 nm (stx2), 568 nm (eae), and 610 nm (internal amplification control pUC18/pUC19).
2.2.3 Module 3: STEC isolation
Strain isolation was carried out as two step procedure. At first, the bacterial suspension from TBX agar was streaked onto TBX agar and CHROMagar STEC (Mast Diagnostica, Reinfeld, Germany, No. 201381) as original suspension, and twice tenfold diluted in 0.9% NaCl resulting in a 10−1 and 10−2 dilution. This resulted in 12 agar plates. Selective agar plates were incubated overnight at 44 °C. If available, ten isolated colonies were selected from each plate and pooled together in 200 µL Aqua bidest; for this purpose approximately half of the bacterial colony was transferred with an inoculation needle. Purple colonies were selected from CHROMagar STEC and blue colonies from TBX, in general. As some E. coli strains, thereunder STEC O157:H7 strains, lack glucuronidase activity, white colonies from TBX agar were selected and pooled the same way. If many different isolated colonies grew on one plate, an area of approximately 0.5 × 0.7 cm from the part with heaviest growth was removed with an inoculation loop and likewise transferred to 200 µL Aqua bidest. DNA was extracted by thermal lysis for 10 min at 95 °C, followed by 30 s centrifugation at 10,000×g, and subjected to multiplex real-time PCR as described. All colonies from positive pools were transferred to nutrient agar (Oxoid, CM0003B). Up to ten colonies were streaked on one agar plate in a star-like pattern. After overnight incubation at 37 °C, colony material from each strain was transferred into 200 µL Aqua bidest and analyzed separately by real-time PCR on a 10 µL scale applying 1 µL bacterial template DNA.
If none of the pools of ten colonies tested positive, but the colony material from the heavy growth part was positive, the second step of the isolation procedure called “500 colony procedure” was started. All bacterial colonies from these plates were transferred with sterile toothpicks to nutrient agar. 20 colonies were streaked onto one nutrient agar in a star-like pattern. By this procedure, on average 500 single colonies (300–700) were transferred from one subsample. After overnight incubation at 37 °C, all colonies were analyzed by real-time PCR as pools per 20 subcultures. Positive pools were separated as described. All strains were verified by the National Reference Laboratory for Escherichia coli, Bundesinstitut für Risikobewertung (BfR) Berlin, Germany (data not shown).
2.3 Interpretation of the results
Samples were reported positive, when STEC was detected and one or more STEC strains were isolated. If the two step isolation procedure did not result in successful strain isolation, but analysis of the part with heavy growth did show the presence of culturable STEC on TBX agar, the subsample was classified as “molecular positive but non isolated”.
2.4 Statistical analysis
Statistical analysis was done using R version 3.2.4 (R Foundation for Statistical Computing). Samples were categorized according to dichotomous variable “positive” or “negative” into k × 2 contingency tables. Contingency tables were analyzed using the Χ2-test. If figures in categories were below 5, the test simulated p value based on 2000 replicates was applied.
2.5 Evaluation of hygienic conditions
On-site inspections of hygienic conditions in mills were carried out during the last 6 years as part of a local monitoring program. Mill H was visited after STEC findings. All inspections were carried together with a company representative in a structured way. Control points included visual checks of equipment, rooms and outside environment, as well documentation checks including records of pest control.
3 Results
Results are summarized in Table 1. STEC was detected in 38 of 98 (39%) test samples by real-time PCR and isolated from 17 test samples out of 88 (19%). Sample 6 with 10 subsamples, of which 7 were positive, was used to confirm the first positive findings (sample 4 and 5). The isolation procedure was not applied to sample 6 resulting in the difference of ten. STEC was successfully isolated from 55% of molecular positive subsamples. STEC could not be isolated from three molecular positive laboratory samples even when the “500 colony procedure” was applied.
Results of molecular detection and strain isolation in combination with the factors E. coli, grain species and mill are summarized in Table 3. Molecular detection of STEC corresponds to the number of E. coli (Χ2 = 7.1018, df = 1, p value = 0.0077). The isolation of STEC was not dependent on the presence of E. coli (Χ2 = 1.1182, df = 1, p value = 0.2903). The grain species did not have an influence on the molecular detection or on the isolation of STEC in flour samples. There was some evidence that STEC detection and strain isolation did cluster at some flourmills, however, statistical significance for this trend could not be found. On-site inspections at mills showed a broad range of hygienic conditions, with a trend towards STEC detection at mills with inferior hygienic conditions.
Incoming samples consisted of 1, 2 or 10 individual subsamples. Two laboratory samples drawn at mill G were composed of two subsamples; these were grouped together with samples from mill H consisting of ten subsamples. As shown in Table 4, detection rate and strain isolation could be improved when laboratory samples consisted of several subsamples.
4 Discussion
Our studies, starting in 2014, showed the presence of STEC in flour in Germany for the 1st time. STEC was detected in wheat and rye flour through a combination of molecular and microbial methods. The presence of at least one of the two genes stx1 and stx2 encoding Shiga toxins is used as the common characteristic for molecular identification of EHEC (Karmali 2004). Molecular detection after two subsequent enrichment steps guarantees positive results by culturable bacteria only. In this work, ISO/TS 13136:2012 (ISO 2012) was modified by the introduction of a 2nd selective enrichment in accordance to other protocols (Tzschoppe et al. 2012; Working Group “Molecular Methods—Microbiology” 2012). There are several advantages of a second specific enrichment step, including better sensitivity, exclusion of the influence of co-extracted food material, and the exclusion of DNA from inactivated cells. The isolation procedure was carried out with a two-step procedure, based on the consideration that isolation rates can be increased with an increase in the number of colonies tested. During the 1st step, approximately 100 colonies were selected and tested as pools of ten each by PCR. In addition to single colonies, material from the part with heavy growth was subjected to molecular analysis. Positive findings there indicated the presence of STEC colonies on this plate. This led to the decision to test as many colonies as possible. Consequently, all colonies from this plate were picked and transferred to nutrient agar plates. This so-called “500 colony procedure” took approximately 3 h per sample which was considered appropriately for this pathogen. Not all isolation attempts were successful; Baumgartner et al. (2016) had comparable results. Despite these difficulties, strain isolation is necessary in official food surveillance as justification for legal actions and for epidemiological studies.
Wheat and rye were grown and harvested under similar conditions in our region and there was no influence of the grain species.
The factor “flourmill” showed a non-significant trend that positive findings occur in some mills only. Grains as raw material are not sterile per se but grains were not contaminated at detectable levels (data not shown). Maintenance and hygienic conditions differ between the mills, e.g. mill H with an ongoing STEC contamination problem suffers from inappropriate hygienic conditions. The milling process includes a conditioning step to adjust the water content at 14–15%. Water is being applied to the grain which is being stored in conditioning cells for several hours afterwards. Berghofer et al. (2003) found a remarkable increase of E. coli counts after conditioning, and detected build-up of grain residues in conditioning augers, elevators and storage bins.
Based on our data, the contamination source could not be found. Two different scenarios could have led to STEC contamination of flour. In the 1st scenario, STEC in contaminated raw material multiplied after the conditioning step in residues adhered to the technical equipment of the mill. In the 2nd scenario, contamination occurred in the mill through contaminated water used for conditioning or by excrement from rodents and birds.
The detection rate increased significantly if the laboratory sample was composed of several subsamples. The higher strain isolation rate with an increasing number of subsamples was not significant. The isolation rate depended on several factors, the number of subsamples was one of these. Our findings might have been biased by the limited number of mills sampled with subsamples. Future programs should define the number of subsamples to allow a proper comparison of analytical results.
Our findings show the frequent presence of STEC in flour in Germany. Future systematic studies over periods of time should analyze the factor “flourmill” in a systematic way, together with the hygienic and technological conditions. Food business operators producing foodstuff containing uncooked flour should be aware of the possibility of the presence of STEC and should consider the higher thermal stability of STEC in products with low water activity (Finn et al. 2013; Liu et al. 2014; Knödler et al. 2016).
References
Baumgartner A, Niederhauser I, Diston D, Moor D (2016) Enterohemorrhagic Escherichia coli (EHEC) in water from karst springs: detection with real-time PCR and isolation of strains. J Consum Prot Food Saf 11:353–357. doi:10.1007/s00003-016-1053-1
Bauwens A, Toval F, Mellmann A, Karch H (2016) Auf der Spur Pflanzen-assoziierter STEC: Kontamination oder Habitat? In: Konrad R (ed) V. EHEC-Workshop 2016 Nördlingen. Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit (LGL) Eggenreuther Weg 43, 91058 Erlangen, Erlangen, pp 30–31
Berghofer LK, Hocking AD, Miskelly D, Jansson E (2003) Microbiology of wheat and flour milling in Australia. Int J Food Microbiol 85:137–149. doi:10.1016/S0168-1605(02)00507-X
Beuchat LR, Komitopoulou E, Beckers H, Betts RP, Bourdichon F, Fanning S, Joosten HM, ter Kuile BH (2013) Low-water activity foods: increased concern as vehicles of foodborne pathogens. J Food Prot 76:150–172. doi:10.4315/0362-028X.JFP-12-211
BVL (1991) Untersuchung von Lebensmitteln—Bestimmung der Anzahl von Hefen und Schimmelpilzen in Milch und Milchprodukten—Referenzverfahren. 1–3. L.00.00-37(1991)
BVL (2014) Berichte zur Lebensmittelsicherheit 2012: Zoonosen-Monitoring 2012. Springer International Publishing AG, Switzerland. doi:10.1007/978-3-319-04409-5
BVL (2015) Berichte zur Lebensmittelsicherheit 2013: Zoonosen-Monitoring 2013. Springer International Publishing AG, Switzerland. doi:10.1007/978-3-319-15380-3
BVL (2016a) Berichte zur Lebensmittelsicherheit 2014: Zoonosen-Monitoring 2014. Springer International Publishing AG, Switzerland. doi:10.1007/978-3-319-30151-8
BVL (2016b) Berichte zur Lebensmittelsicherheit 2015: Zoonosen-Monitoring 2015. http://www.bvl.bund.de/SharedDocs/Downloads/01_Lebensmittel/04_Zoonosen_Monitoring/Zoonosen_Monitoring_Bericht_2015.pdf?__blob=publicationFile&v=6. Accessed 17 Mar 2017
Byrd-Bredbenner C, Abbot JM, Wheatley V, Schaffner D, Bruhn C, Blalock L (2008) Risky eating behaviors of young adults—implications for food safety education. J Am Diet Assoc 108:549–552. doi:10.1016/j.jada.2007.12.013
CDC (2016) Multistate outbreak of Shiga toxin-producing Escherichia coli O121 infections linked to flour. In: Centers Dis. Control Prev. 24/7 Sav. Lives, Prot. people. http://www.cdc.gov/ecoli/2016/o121-06-16/index.html. Accessed 26 Feb 2017
Clark CW, Ordal ZJ (1969) Thermal injury and recovery of Salmonella typhimurium and its effect on enumeration procedures. Appl Microbiol 18:332–336
EFSA Panel on Biological Hazrads (BIOHAZ) (2013) Scientific opinion on VTEC-seropathotype and scientific criteria regarding pathogenicity assessment. EFSA J 11(4):1–106. doi:10.2903/j.efsa.2013.3138
Finn S, Condell O, McClure P, Amézquita A, Fanning S (2013) Mechanisms of survival, responses and sources of Salmonella in low-moisture environments. Front. Microbiol. 4:331
Gieraltowski L, Schwensohn C, Meyer S, Eikmeier D, Medus C, Sorenson A, Forstner M, Madad A, Blankenship J, Feng P, Williams I (2017) Multistate outbreak of Escherichia coli O157:H7 infections linked to dough mix—United States, 2016. Morb Mortal Wkly Rep 66:88–89. doi:10.15585/mmwr.mm6603a6
Gilbert S, Lake R, Cressey P, King N (2010) Risk profile: Salmonella (non typhoidal) in cereal grains. http://www.foodsafety.govt.nz/elibrary/industry/salmonella-in-cereals.pdf. Accessed 17 Mar 2017
Greene SEL (2012) Thermal inactivation of Escherichia coli O157: H7 and Salmonella Agona in Wheat Flour. Thesis. Virginia Polytechnic Institute and State University
Hoorfar J, Baggesen DL (1998) Importance of pre-enrichment media for isolation of Salmonella spp. from swine and poultry. FEMS Microbiol Lett 169:125–130. doi:10.1016/S0378-1097(98)00474-1
ISO (2001) ISO 16649-2:2001 Microbiology of food and animal feeding stuffs—horizontal method for the enumeration of beta-glucuronidase-positive Escherichia coli—part 2: colony-count technique at 44 degrees C using 5-bromo-4-chloro-3-indolyl beta-d-glucuronide
ISO (2003) ISO 6888-1:1999/Amd 1:2003 Microbiology of food and animal feeding stuffs—horizontal method for the enumeration of coagulase-positive staphylococci (Staphylococcus aureus and other species)—part 1: technique using Baird-Parker agar medium
ISO (2004) ISO 7932:2004 Microbiology of food and animal feeding stuffs—horizontal method for the enumeration of presumptive Bacillus cereus—Colony-count technique at 30 degrees C
ISO (2012) ISO/TS 13136:2012 Microbiology of food and animal feed—real-time polymerase chain reaction (PCR)-based method for the detection of food-borne pathogens—horizontal method for the detection of Shiga toxin-producing Escherichia coli (STEC) and the determination of O157, O111, O26, O103 and O145 serogroups
ISO (2013) ISO 4833-1:2013 Microbiology of the food chain—horizontal method for the enumeration of microorganisms—part 1: colony count at 30 degrees C by the pour plate technique
Karch H, Leopold SR, Kossow A, Mellmann A, Köck R, Bauwens A (2015) Enterohemorrhagic E. coli (EHEC): environmental-vehicle-human interface. In: Sing A (ed) Zoonoses—infections affecting humans and animals: focus on public health aspects. Springer, Dordrecht, pp 235–248
Karmali MA (2004) Prospects for preventing serious systemic toxemic complications of Shiga toxin-producing Escherichia coli infections using Shiga toxin receptor analogues. J Infect Dis 189:355–359. doi:10.1086/381130
Knödler M, Berger M, Dobrindt U (2016) Long-term survival of the Shiga toxin-producing Escherichia coli O104: H4 outbreak strain on fenugreek seeds. Food Microbiol 59:190–195. doi:10.1016/j.fm.2016.06.005
Liu S, Anderson NM, Fleischman GJ, Grasso EM (2014) Thermal resistance study of STEC in low moisture foods using differential scanning calorimeter (DSC). https://www.ifsh.iit.edu/system/files/members/resources/grasso-ecoli.pdf. Accessed 17 Mar 2017
Mäde D, Reiting R, Strauch E, Ketteritzsch K, Wicke A (2008) A real-time PCR for Detection of Pathogenic Yersinia enterocolitica in food combined with an Universal Internal Amplification Control System. J Consum Prot Food Saf 3:141–151. doi:10.1007/s00003-008-0341-9
Neil KP, Biggerstaff G, Macdonald JK, Trees E, Medus C, Musser KA, Stroika SG, Zink D, Sotir MJ (2012) A novel vehicle for transmission of Escherichia coli O157: H7 to humans: multistate outbreak of E. coli O157: H7 infections associated with consumption of ready-to-bake commercial prepackaged cookie Dough—United States, 2009. Clin Infect Dis 54:511–518. doi:10.1093/cid/cir831
Nielsen EM, Andersen MT (2003) Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J Clin Microbiol 41:2884–2893. doi:10.1128/JCM.41.7.2884
Pavlovic M, Huber I, Skala H, Konrad R, Schmidt H, Sing A, Busch U (2010) Development of a multiplex real-time polymerase chain reaction for simultaneous detection of enterohemorrhagic Escherichia coli and enteropathogenic Escherichia coli strains. Foodborne Pathog Dis 7:801–808. doi:10.1089/fpd.2009.0457
Perelle S, Dilasser F, Grout J, Fach P (2004) Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world’s most frequent clinical cases. Mol Cell Probes 18:185–192. doi:10.1016/j.mcp.2003.12.004
Reischl U, Youssef MT, Kilwinski J, Lehn N, Zhang WL, Karch H, Strockbine NA (2002) Real-time fluorescence PCR assays for detection and characterization of Shiga Toxin, intimin, and enterohemolysin genes from Shiga Toxin-producing Escherichia coli. J Clin Microbiol 40:2555–2565. doi:10.1128/JCM.40.7.2555
Robert Koch Institut (2011) Report: Final presentation and evaluation of epidemiological findings in the EHEC O104:H4 Outbreak Germany 2011. http://edoc.rki.de/documents/rki_ab/reQHS31jDrGxc/PDF/23NXL3JomOyAA.pdf. Accessed 17 Mar 2017
Tzschoppe M, Martin A, Beutin L (2012) A rapid procedure for the detection and isolation of enterohaemorrhagic Escherichia coli (EHEC) serogroup O26, O103, O111, O118, O121, O145 and O157 strains and the aggregative EHEC O104:H4 strain from ready-to-eat vegetables. Int J Food Microbiol 152:19–30. doi:10.1016/j.ijfoodmicro.2011.10.009
Working Group “Molecular Methods—Microbiology” (2012) Ringversuchsvorschrift: Nachweises von Shiga-Toxin bildenden Escherichia coli (STEC) in frischen pflanzlichen Lebensmitteln Real-time PCR-Verfahren. Working document Berlin 2012
Wu S, Ricke SC, Schneider KR, Ahn S (2017) Food safety hazards associated with ready-to-bake cookie dough and its ingredients. Food Control 73, Part B:986–993. doi:10.1016/j.foodcont.2016.10.010
Acknowledgements
We thank Christine Degner, Ilona Balogh, Katja Trübner-Mäde, Birgit Jahn, Christiane Klemm, and Juliane Voigt for the excellent technical assistance. We would like to thank John Church for carefully reading the manuscript and for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mäde, D., Geuthner, AC., Imming, R. et al. Detection and isolation of Shiga-Toxin producing Escherichia coli in flour in Germany between 2014 and 2017. J Consum Prot Food Saf 12, 245–253 (2017). https://doi.org/10.1007/s00003-017-1113-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00003-017-1113-1