Abstract
The classification of two root-infecting fungi, Magnaporthe garrettii and M. griffinii, was examined by phylogenetic analysis of multiple gene sequences. This analysis demonstrated that M. garrettii and M. griffinii were sister species that formed a well-supported separate clade in Papulosaceae (Diaporthomycetidae, Sordariomycetes), which clusters outside of the Magnaporthales. Wongia gen. nov, is established to accommodate these two species which are not closely related to other species classified in Magnaporthe nor to other genera, including Nakataea, Magnaporthiopsis and Pyricularia, which all now contain other species once classified in Magnaporthe.
Similar content being viewed by others
Introduction
The taxonomic and nomenclatural problems that surround generic names in the Magnaporthales (Sordariomycetes, Ascomycota), together with recommendations for the suppression and protection of some of these names, were explained by the Pyricularia/Magnaporthe Working Group established under the auspices of the International Commission on the Taxonomy of Fungi (ICTF; Zhang et al. 2016). One of these generic names, Magnaporthe, was proposed for suppression by Zhang et al. (2016) because Magnaporthe is congeneric with Nakataea (Hara 1939) as the types of both genera, Magnaporthe salvinii (syn. Leptosphaeria salvinii) and Nakataea sigmoidea (syn. Helminthosporium sigmoideum) are conspecific(Krause & Webster 1972, Luo & Zhang 2013).
Magnaporthe was morphologically characterised by having dark perithecia with long necks immersed in host tissue, unitunicate asci, and 4-celled fusiform hyaline to pale brown ascospores (Krause & Webster 1972). Subsequently, seven species were assigned to Magnaporthe based on morphology, namely, M. salvinii (Krause & Webster 1972), M. grisea (Barr 1977), M. rhizophila (Scott & Deacon 1983), M. poae (Landschoot & Jackson 1989), M. oryzae (Couch & Kohn 2002), and M. garrettii and M. griffinii (Wong et al. 2012). Most of these species belong to other genera, specifically Magnaporthiopsis, Nakataea, and Pyricularia (Luo & Zhang 2013). The two exceptions are the Australian ectotrophic species, M. garrettii and M. griffinii, which infect roots of some turf grasses (Wong et al. 2012). One of these species, M. griffinii, was found by Klaubauf et al. (2014) to be distant from Sordariomycetes based on ITS sequences (GenBank JQ390311, JQ390312).
This study aims to resolve the classification of M. garrettii and M. griffinii using molecular sequence data from the type specimens. Four loci from the nuclear genome namely, ITS) and the large subunit (LSU) of rDNA, translation elongation factor 1-alpha (TEF1), and the largest subunit of RNA polymerase II (RPB1) were selected for analysis.
Materials and Methods
Fungal cultures and DNA extraction
Dried specimens of the holotypes of Magnaporthe garrettii (DAR 76937) and M. griffinii (DAR 80512) were borrowed from the Plant Pathology Herbarium, New South Wales Agriculture (DAR). Dried perithecia were excised with a needle and soaked in extraction buffer overnight at 65 °C before extraction of DNA with an UltraClean® Microbial DNA Isolation Kit (MoBIO Laboratories) as per the manufacturer’s instructions. An additional culture of M. griffinii (BRIP 60377) was grown on PDA for 6 wk before enough mycelium was produced for DNA extraction.
PCR amplification
The primer pairs ITS1/ITS4 (White et al. 1990), RPB-Ac/RPB-Cr (Castlebury et al. 2004, Matheny et al. 2002), LR5/LROR and EF1983F/2218R (Schoch et al. 2009) were used to amplify ITS, RPB1, LSU, and TEF1 sequences, respectively. PCR amplifications were conducted in a 20 µl reaction volume containing 1 µl of 5–10 ng DNA, 10 µl of high fidelity Phusion DNA Polymerase (New England Biolabs), 1 µl of primers (10 µM) and 7 µl of sterile water with the thermal cycling program as follows: 98 °C for 30s, 30 cycles of 98 °C for 10 s, 58–62 °C for 30 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min. PCR products were sent to Macrogen (Korea) for direct sequencing using the amplification primers.
Phylogenetic analysis
All sequences were assembled with Sequencher v. 5.1 (Gene Codes, Ann Arbor, MI). Alignments were generated for individual loci using MAFFT v. 6.611 (Katoh & Toh 2008), and then the alignments concatenated for the phylogenetic analyses. DNA sequences were deposited in GenBank with the accession numbers listed in Table 1 and the final curated alignment deposited in TreeBASE under accession no. ID 19968. Phylogenetic trees were reconstructed with two phylogenetic criteria, Maximum likelihood (ML) and Bayesian Inference (BI). ML was carried out with RAxML v. 7.2.6 using GTRGAMMA as the model of evolution (Stamatakis 2006), choosing the rapid bootstrap analysis (command —f a) with a random starting tree and 1000 maximum likelihood bootstrap replications. BI was done with MrBayes v. 3.1.2 (Ronquist et al. 2012), utilizing four parallel MCMC chains, which were allowed to run for 10 million generations, with sampling every 1000 generations and saving trees every 5 000 generations. The cold chain was heated at a temperature of 0.25. All phylogenetic trees were visualized using FigTree (Morariu et al. 2009).
Results
Molecular phylogeny
The phylogenetic trees recovered from the ML and BI analyses had identical topologies and were well-supported by bootstrap and posterior probabilities (Fig. 1). The analyses comprised 36 taxa belonging to eight orders and two families in the subclass Diaporthomycetidae (Sordariomycetes). Camarops ustulinoides (Boniliales, Sordariomycetes) was used as the outgroup (Table 1). The phylogenetic analysis revealed Magnaporthe garrettii (DAR 76937) and M. griffinii (DAR 80512) as sister species that formed a distinct well-supported (100/1.0) monophyletic clade in Papulosaceae that sat outside Magnaporthales. The analysis provided moderate support (67/0.93) for placement of M. garrettii and M. griffinii in Papulosaceae, which has not yet been assigned to any order of Diaporthomycetidae. Based on this analysis, a new generic name is established here to accommodate M. garrettii and M. griffinii.
Phylogenetic tree obtained from a maximum likelihood analysis of the combined ITS/LSU/RPB1/TEF1 alignment. The bootstrap support values from 1 000 replicates and posterior probabilities obtained in Bayesian analysis are indicated at the nodes. The scale bar indicates the expected changes per site. Ex-type cultures of species are indicated in bold.
Taxonomy
Wongia Khemmuk, Geering & R.G. Shivas, gen. nov.
MycoBank MB817529
Etymology: Named after the eminent Australian mycologist and plant pathologist, Percy T.W. Wong (University of Sydney), who first studied and classified these fungi.
Diagnosis: Differs from all other genera in the subclass Diaporthomycetidae in having non-amyloid apical rings in the asci with 3-septate ascospores that have dark brown middle cells and pale brown to subhyaline shorter distal cells.
Type species: Wongia garrettii (P. Wong & M.L. Dickinson) Khemmuk et al. 2016
Classification: Ascomycota, Sordariomycetes, Diaporthomycetidae.
Description: Mycelium comprised of brown, straight or flexuous hyphae, with simple hyphopodia. Ascomata perithecial, superficial and immersed, mostly solitary or sometimes aggregated in small groups, globose, black, ostiolate, with a long or short neck, perithecial wall composed of textura epidermoidea, external cell much darker. Paraphyses thin-walled, hyaline, filiform, septate.
Asci unitunicate in structure, cylindrical, mostly straight, short stalked, tapered towards a rounded apex, with a light refractive, non-amyloid apical ring, 8-spored. Ascospores uniseriate, cylindrical to fusiform, straight or slightly curved with rounded ends, 3-septate, middle cells dark brown and distal cells pale brown to subhyaline and shorter.
Wongia garrettii (P. Wong & M.L. Dickinson) Khemmuk, Geering & R.G. Shivas, comb. nov.
MycoBank MB817530
Basionym: Magnaporthe garrettii P. Wong & M. L. Dickinson, Australasian Plant Pathology 41: 326 (2012).
Type: Australia: South Australia: Adelaide, Colonel Light Gardens Bowling Club, on Cynodon dactylon, 30 Oct. 2004, M.L. Dickinson (DAR 76937 — holotype).
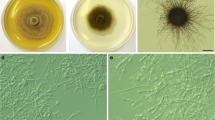
Description and illustration: Wong et al. (2012).
Wongia griffinii (P.Wong & A.M. Stirling) Khemmuk, Geering & R.G. Shivas, comb. nov.
MycoBank MB817531
Basionym: Magnaporthe griffinii P. Wong & A.M. Stirling, Australasian Plant Pathology 41: 327 (2012).
Type: Australia: Queensland: Coolum, Hyatt Coolum Golf Club, on Cynodon dactylon × transvaalensis, 13 Mar. 2008, M. Whatman (DAR 80512 — holotype).
Description and illustration: Wong et al. (2012)
Other specimens examined: Australia: New South Wales: Cobbitty, on Cynodon dactylon, 19 Apr. 2013, G. Beehag, (BRIP 60378). Queensland: Brisbane, on on Cynodon dactylon × transvaalensis, Jan. 2000, A.M. Stirling (BRIP 60377).
Discussion
Magnaporthe is a synonym of Nakataea as their respective type species, Magnaporthe salvinii and Nakataea sigmoidea, refer to the same species (Krause & Webster 1972, Luo & Zhang 2013, Klaubauf et al. 2014, Zhang et al. 2016). This led us to re-examine two Australian species, M. garrettii and M. griffinii, pathogenic on roots of couch (Cynodon dactylon) and hybrid couch (C. dactylon × transvaalensis) (Wong et al. 2012). We establish Wongia here to accommodate these two species, based on molecular and morphological analysis.
Multigene analyses placed W. garrettii and W. griffinii in Papulosaceae (Diaporthomycetidae, Sordariomycetes; Maharachchikumbura et al. 2015) with moderate bootstrap support (Fig. 1). The Papulosaceae has not yet been placed in an order within Sordariomycetes (Winka & Erikson 2000). Wongia is the fourth genus to be placed in Papulosaceae, along with Brunneosporella (Ranghoo & Hyde 2001), Fluminicola (Wong et al. 1999). and Papulosa (Kohlmeyer & Volkmann-Kohlmeyer 1993). Most members in this family are found on submerged wood in freshwater habitats and grow slowly in culture on potato dextrose agar (Ranghoo & Hyde 2001). Wongia garrettii and W. grifinii are morphologically different from other genera of Papulosaceae in having non-amyloid apical rings in the asci using Melzer’s reagent, while others have amyloid apical rings (Winka & Eriksson (2000). The long perithecial necks of W. garrettii differentiate it from W. griffinii (Wong et al. 2012), which also has larger ascospores (24–35 × 6–9 µm) than W. garrettii (19–25 × 5–7 µm) (Wong et al. 2012). Asexual morphs have not been found in either W. garrettii or W. griffinii in nature or in cultures grown on artificial media under laboratory conditions (Wong et al. 2012).
References
Barr ME (1977) Magnaporthe, Telimenella, and Hyponectria (Physosporellaceae). Mycologia 69: 952–966.
Castlebury LA, Rossman AY, Sung GH, Hyten AS, Spatafora JW (2004) Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycological Research 108: 864–872.
Couch BC, Kohn LM (2002) A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94: 683–693.
Hara K (1939) The Diseases of the Rice-plant. 2nd edn. Gifu: Japanese Society for Fungi.
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298.
Klaubauf S, Tharreau D, Fournier, E, Groenewald JZ, Crous PW, et al. (2014) Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Studies in Mycology 79: 85–120.
Kohlmeyer J, Volkmann-Kohlmeyer B (1993) Two new genera of Ascomycotina from saltmarsh Juncus. Systema Ascomycetum 11: 95–106.
Krause RA, Webster RK (1972) The morphology, taxonomy, and sexuality of the rice stem rot fungus, Magnaporthe salvinii (Leptosphaeria salvinii). Mycologia 64: 103–114.
Landschoot PJ, Jackson N (1989) Magnaporthe poae sp. nov., a hyphopodiate fungus with a Phialophora anamorph from grass roots in the United States. Mycological Research 93: 59–62.
Luo J, Zhang N (2013) Magnaporthiopsis, a new genus in Magnaporthaceae (Ascomycota). Mycologia 105: 1019–1029.
Maharachchikumbura SN, Hyde K, Jones EBG, McKenzie EC, Huang SK, et al. (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity 72: 199–301.
Matheny PB, Liu YJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany 89: 688–698.
Morariu VI, Srinivasan BV, Raykar VC, Duraiswami R, Davis LS (2009) Automatic online tuning for fast Gaussian summation. In: Advances in Neural Information Processing Systems 21: 22nd Annual Conference on Neural Information Processing Systems 2008 (Proceedings of a meeting held 8-10-December 2008, Vancouver) (Koller D, Schuurmans D, Bengio Y, Bottou L, eds): 1113–1120. Neural Information Processing Systems.
Ranghoo VM, Tsui CKM, Hyde KD (2001) Brunneosporella aquatica gen. et sp. nov., Aqualignicola hyalina gen. et sp. nov., Jobellisia viridifusca sp. nov. and Porosphaerellopsis bipolaris sp. nov. (ascomycetes) from submerged wood in freshwater habitats. Mycological Research 105: 625–633.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Schoch CL, Crous PW, Groenewald JZ, Boehm EWA, Burgess TI, et al. (2009) A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15.
Scott DB, Deacon JW (1983) Magnaporthe rhizophila sp. nov., a dark mycelial fungus with a Phialophora conidial state, from cereal roots in South Africa. Transactions of the British Mycological Society 81: 77–81.
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis M, Gelfand D, Shinsky J, White T, eds): 315–322. San Diego: Academic Press.
Winka K, Eriksson OE (2000) Papulosa amerospora accommodated in a new family (Papulosaceae, Sordariomycetes, Ascomycota) inferred from morphological and molecular data. Mycoscience 41: 97–103.
Wong PTW, Dong C, Stirling AM, Dickinson ML (2012) Two new Magnaporthe species pathogenic to warm-season turf grasses in Australia. Australasian Plant Pathology 41: 321–329.
Wong SW, Hyde KD, Jones EBG (1999) Ultrastructural studies on freshwater ascomycetes, Fluminicola bipolaris gen. et sp. novo. Fungal Diversity 2: 189–197.
Zhang N, Luo J, Rossman AY, Aoki T, Chuma I, et al. (2016) Generic names in Magnaporthales. IMA Fungus 7: 155–159.
Acknowledgements
We acknowledge support of the Australian Government’s Cooperative Research Centres Program (project no. PBCRC62082), and the Plant Pathology Herbarium, New South Wales Agriculture (DAR), for lending us the type specimens of the two species for sequencing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Khemmuk, W., Geering, A.D.W. & Shivas, R.G. Wongia gen. nov. (Papulosaceae, Sordariomycetes), a new generic name for two root-infecting fungi from Australia. IMA Fungus 7, 247–252 (2016). https://doi.org/10.5598/imafungus.2016.07.02.04
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.5598/imafungus.2016.07.02.04