Abstract
The recently described fungal phylum Entorrhizomycota was established solely for the genus Entorrhiza, species of which cause root-galls in Cyperaceae and Juncaceae. Talbotiomyces calosporus (incertae sedis) shares morphological characteristics and an ecological niche with species of Entorrhiza. We investigated the higher classification of T. calosporus to determine whether it belongs in Entorrhizomycota. Ribosomal DNA sequences showed Talbotiomyces to be a close relative of Entorrhiza and both taxa form a highly supported monophyletic group. Based on molecular phylogenetic analyses and in congruence with existing morphological and ecological data, Entorrhiza and Talbotiomyces represent a deep dichotomy within the Entorrhizomycota. While species of Entorrhiza are characterised by dolipores and occur on monocotyledons, members of Talbotiomyces are characterised by simple pores and are associated with eudicotyledons. This expands the host range of the recently described Entorrhizomycota from Poales to other angiosperms. Higher taxa, namely Talbotiomycet ales ord. nov. and Talbotiomycetaceae fam. nov., are proposed here to accommodate Talbotiomyces.
Similar content being viewed by others
Introduction
Entorrhiza consists of species that form galls on roots of two monocotyledon sister families, Cyperaceae and Juncaceae. This enigmatic genus had an unstable and unresolved systematic position within the kingdom Fungi. Traditionally, it was considered a smut genus and classified in Tilletiaceae (Zundel 1953). In the modern classification of smut fungi, based on ultrastructure and molecular phylogeny, Entorrhiza was accommodated in the distinct family, order, subclass and class, Entorrhizaceae, Entorrhizales, Entorrhizomycetidae and Entorrhizomycetes, within the Ustilaginomycotina (Bauer et al. 1997, Begerow et al. 1998, 2007). Bauer et al. (2015) demonstrated that Entorrhiza could not be assigned to any of the main fungal lineages and consequently established the novel phylum Entorrhizomycota. However, it remains unclear if Entorrhizomycota belongs to the subkingdom Dikarya, which comprises the phyla Ascomycota and Basidiomycota, or represents a sister group (Bauer et al. 2015).
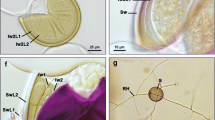
Entorrhizomycota currently comprises a single genus, Entorrhiza, with 14 species that cause galls in the roots of Cyperaceae and Juncaceae (Fineran 1978, Vánky 1994, 2012, 2013, Denchev & Denchev 2012). Due to the different age estimates of the Entorrhizomycota versus those of their host groups, Bauer et al. (2015) hypothesized that known entorrhizomycotan species and their respective host range only reflect a very small portion of a much larger, yet undiscovered diversity. Species of Entorrhiza regularly form septate, clampless hyphae, which are coiled in living host cells. During spore formation, hyphae terminate with globose cells, which detach from the hyphae and become thick-walled teliospores (Fineran 1980, Deml & Oberwinkler 1981, Bauer et al. 2015). Talbotiomyces, a genus morphologically similar to Entorrhiza, was established to accommodate Entorrhiza calospora, a species causing galls on roots of some South African members of the eudicotyledonous order Caryophyllales (Vánky et al. 2007). Given the similar morphology and ecology of Talbotiomyces calosporus to those of Entorrhiza species, it is possible that Talbotiomyces might be an unrecognized member of the Entorrhizomycota. To test this hypothesis, we analysed the nuclear rDNA of an isotype of T. calosporus collected in 1943, with five species of Entorrhiza, and representatives of Dikarya and Glomeromycota.
Materials and Methods
Material
Specimen examined: Talbotiomyces calosporus: South Africa: Gauteng: Pretoria, Brummeria, near Murray Farm, on Limeum viscosum (syn. L. glomeratum), 30 Jan. 1943, J.J.O. Pazzi (BRIP: HUV 587 — isotype from PREM 33770 — holotype).
DNA isolation, PCR, and sequencing
Total genomic DNA was extracted using the InnuPREP Plant DNA Kit (Analytik Jena, Jena) according to the manufacturer’s instructions. The small subunit (SSU) rDNA region was amplified with various primer combinations from the sets NS1 to NS8 (White et al. 1990) and NS17 to NS24 (Gargas & Taylor 1992). PCRs were performed using Phusion High-Fidelity DNA Polymerase (Finnzymes Oy, Vantaa, Finland), following the protocol recommended by the manufacturer with annealing temperatures of approximately 5 °C above the mean primer melting temperatures, and 30 cycles. The 5′ end of the large subunit (LSU) of the nuclear rDNA was amplified with the primer combination LR0R (R. Vilgalys Lab., http://biology.duke.edu/fungi/mycolab/primers.htm) and LR6 (Vilgalys & Hester 1990). PCR mixtures consisted of 5 µL of PCR reaction buffer (10×), 0.75 mM of MgCl2 (50 mM), 14.5 µL of water, 1 µL of dNTP mix (5 mM), 0.50 µL of each primer (25 pmol/µL), 0.25 µL of MangoTaq DNA polymerase (Bioline, Luckenwalde) (2 U/µL), and undiluted 2.5 µL template DNA. PCRs were performed with the following thermal profile: 10 cycles of 30 s at 94 °C, 45 s at 60 °C (−1 °C per cycle), and 75 s at 72 °C, followed by 26 cycles of 30 s at 94 °C, 45 s at 50 °C and 75 s at 72 °C, with a final extension of 7 min at 72 °C. PCR products were purified using ExoSAP-IT (USB Corporation, Cleveland, OH), and diluted (1:20) for further analysis. Cycle sequencing of SSU and LSU rDNA was accomplished using a 1:6 diluted BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and the amplification primers. Sequencing was performed on an ABI Prism 3130xl Genetic Analyser (Applied Biosystems). Forward and reverse sequence chromatograms were checked for accuracy and edited using Sequencher v. 4.1 (Gene Codes Corporation, Ann Arbor, MI). The newly generated DNA sequences of Talbotiomyces calosporus have been deposited in GenBank under the accession numbers KJ666129 (SSU) and KP413056 (LSU).
Molecular phylogenetic analyses
To determine the phylogenetic position of Talbotiomyces, we assembled datasets containing rDNA SSU and LSU sequences. Both datasets included a sequence of the isotype of T. calosporus, all Entorrhiza sequences currently available in GenBank and two representatives of each dikaryan subphylum and the Glomeromycota (Fig. 1). The chytrids Gromochytrium mamkaevae and Olpidium brassicae were used as outgroup taxa. For GenBank accession numbers and full datasets see TreeBASE submission ID 17604. Alignments were performed separately with MAFFT v. 7.147b (Katoh & Standley 2013) using the E-INS-i option (Katoh et al. 2005). Initial alignments were further modified with Gblocks v. 0.91b (Castresana 2000) to remove ambiguously aligned regions using standard parameters with one exception: ‘allowed gap positions with half’. The final SSU alignment had a length of 1498 bp and the LSU alignment had a length 514 bp. For further analyses, both alignments were analysed separately as well as in a concatenated dataset. Maximum likelihood (ML) phylogenetic analyses and rapid bootstrapping with 1000 replicates under the GTRCAT model were performed using RAxML v. 8.0.17 (Stamatakis 2014). Additionally, posterior probabilities for nodal support were determined in a Bayesian phylogenetic MCMC search with MrBayes v. 3.2.2 (Ronquist et al. 2012) using the GTR+G model. Each search comprised two runs of four chains, each for 5 × 106 generations. Posterior probabilities were sampled every 100 generations with the first 2.5 × 106 generations being discarded as burn-in. The genetic divergences (uncorrected p-distance) between Talbotiomyces and Entorrhiza were calculated based on full-length sequence alignments using Mesquite v. 2.75 (Maddison & Maddison 2011).
Phylogenetic placement of Talbotiomyces calosporus. Best ML tree topology derived from a combined dataset of SSU and LSU rDNA sequences of Talbotiomyces, Entorrhiza, and representatives of Ascomycota, Basidiomycota, and Glomeromycota. Branch support is given as ML bootstrap percentage (≥ 70) / Bayesian posterior probability (≥ 0.90). The tree was rooted with the chytrids Gromochytrium mamkaevae and Olpidium brassicae.
Results
Phylogenetic placement of Talbotiomyces
The results of our molecular phylogenetic analyses of rDNA sequences of Talbotiomyces calosporus, five species of Entorrhiza, and two representatives of each dikaryan subphylum and Glomeromycota are shown in Fig. 1. In phylogenetic analyses of individual SSU and LSU data (not shown) as well as the combined dataset (Fig. 1), Entorrhiza and Talbotiomyces formed a strongly supported monophyletic group. The genetic distance between T. calosporus and the closest related species was 77 bp or 5.0% to E. fineranae (18S) and 57 bp or 9.0% to E. aschersoniana (LSU). In the phylogenetic tree derived from a concatenated dataset, the Entorrhiza/Talbotiomyces clade was placed as a sister to the Dikarya, supported by 99% ML bootstrap and 1.00 Bayesian posteriorprobability (Fig. 1).
Taxonomy
Our results indicate that Talbotiomyces calosporus belongs to Entorrhizomycota. Molecular phylogenetic analyses revealed a dichotomy between Entorrhiza and Talbotiomyces, and together with ultrastructural and host range information, it justifies a new classification at a higher taxonomical level. We assume a much larger potential diversity of Talbotiomyces, and propose to accommodate Talbotiomyces in a new family and order within the class Entorrhizomycetes:
Talbotiomycetales K. Riess, R. Bauer, R. Kellner, Kemler, Piątek, Vánky & Begerow, ord. nov. MycoBank MB810797
Diagnosis: Members of the Entorrhizomycetes Begerow et al. 2007 having simple pores.
Type: Talbotiomyces Vánky et al. 2007.
Talbotiomycetaceae K. Riess, R. Bauer, R. Kellner, Kemler, Piątek, Vánky & Begerow, fam. nov. MycoBank MB810798
Diagnosis: Members of the Talbotiomycetales occurring on eudicotyledons.
Type: Talbotiomyces Vánky et al. 2007.
Discussion
Entorrhiza/Talbotiomyces relationship
Talbotiomyces calosporus was recovered in a monophyletic group with species of Entorrhiza, with strong phylogenetic support from the SSU and LSU regions of rDNA (Fig. 1). The phylogenetic relationship between Talbotiomyces and Entorrhiza is additionally well supported by morphological and ecological characters of both genera. Both cause galls on plant roots and regularly have septate and clampless hyphae, which form coils in living host cells. Entorrhiza and Talbotiomyces also have hyphae that terminate with globose cells that detach from the hyphae and become thick-walled teliospores (Talbot 1956, Fineran 1980, Deml & Oberwinkler 1981, Bauer et al. 1997, 2001, 2015, Vánky et al. 2007). However, the septal pore architecture and host range of Entorrhiza differs from Talbotiomyces. Entorrhiza is characterised by dolipores and colonizes monocotyledons (Cyperaceae and Juncaceae) (Bauer et al. 1997, 2001). In contrast, T. calosporus possesses simple pores with rounded pore lips and occurs on members of the dicotyledonous order Caryophyllales (Vánky et al. 2007). Therefore, multiple lines of evidence indicate that Entorrhizomycota should be split into two orders: Entorrhizales and Talbotiomycetales, the latter name is introduced in this study. The large genetic distance between Talbotiomyces and Entorrhiza indicates an early divergence of the Entorrhizomycota into two lineages. The inclusion of Talbotiomyces into Entorrhizomycota, however, did not resolve the phylogenetic position of this recently established fungal phylum (Bauer et al. 2015).
Coevolution and presumed host spectrum
Specimens of Entorrhiza and Talbotiomyces are rarely collected and their evolutionary relationships are still poorly understood. Entorrhiza species are associated with roots of Cyperaceae and Juncaceae worldwide, although mostly reported from temperate or high mountain regions (Vánky 2012). Talbotiomyces calosporus is only known from a few localities in subtropical South Africa on Limeum viscosum (Molluginaceae) and Trianthema pentandra (Aizoaceae) (Talbot 1956). Furthermore, Vánky et al. (2007) reported an additional host plant, Portulaca oleracea (Portulacaceae), apparently based on the material preserved in HUV 21397. That specimen is annotated by K. Vánky: “I was unable to find any spores of Entorrhiza calospora type”. The T calosporus/ P. oleracea fungus/host combination needs to be confirmed. It is unclear whether T calosporus is a polyphagous species that associates with host plants from two or three different families, or represents a suite of closely related cryptic or pseudo-cryptic species, each restricted to one host genus or species, as in other biotrophic plant pathogens (e.g. Bauer et al. 2008, Lutz et al. 2008, Göker et al. 2009, Ploch et al. 2011, Beenken et al. 2012, Liu and Hambleton 2013, Piątek et al. 2012, 2013, Vasighzadeh et al. 2014, Voglmayr et al. 2014, McTaggart et al. 2015). Due to the different age estimates of the Entorrhizomycota and their hosts, Bauer et al. (2015) hypothesized that the known species and their respective host range reflects only a small part of a larger diversity. The inclusion of Talbotiomyces in Entorrhizomycota extends the host spectrum of the phylum from Cyperaceae/Juncaceae (Poales) to the eudicotyledons. A large gap remains between the age estimates of Entorrhizomycota (roughly 600 Mya; Bauer et al. 2015) and their hosts (roughly 150 Mya; Wikström et al. 2001). The possibility that the host spectrum of Entorrhizomycota is even broader, comprising also early-diverged land plants, such as ferns and mosses. Due to the lack of above-ground symptoms of infection, infected host plants of Entorrhizomycota are difficult to detect in nature, and such evolutionarily ancient interactions have not been discovered so far. Additionally, some members of the Entorrhiza/ Talbotiomyces clade may not cause symptoms or may occur as endophytes in roots. The next step will be to develop specific primers for Entorrhizomycota to test whether species of this fascinating group of root-colonizing fungi also may occur as endophytes in plant roots, in analogy to Sebacinales (Weiß et al. 2011, Garnica et al. 2013).
References
Bauer R, Begerow D, Oberwinkler F, Piepenbring M, Berbee ML (2001) Ustilaginomycetes. In: The Mycota. Vol. 7. Part B. Systematics and Evolution. (McLaughlin DJ, McLaughlin EG, Lemke PA, eds): 57–83. Berlin: Springer.
Bauer R, Garnica S, Oberwinkler F, Riess K, Weiß M, et al. (2015) Entorrhizomycota: a new fungal phylum reveals new perspectives on the evolution of Fungi. PLoS ONE: in press.
Bauer R, Lutz M, Begerow D, Piatek M, Vánky K, et al. (2008) Anther smut fungi on monocots. Mycological Research 112: 1297–1306.
Bauer R, Oberwinkler F, Vánky K (1997) Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany 75: 1273–1314.
Beenken L, Zoller S, Berndt R (2012) Rust fungi on Annonaceae II: the genus Dasyspora Berk. & M.A. Curtis. Mycologia 104: 659–681.
Begerow D, Bauer R, Oberwinkler F (1998) [“1997”] Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Canadian Journal of Botany 75: 2045–2056.
Begerow D, Stoll M, Bauer R (2007) [“2006”] A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98: 906–916.
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552.
Deml G, Oberwinkler F (1981) Studies in heterobasidiomycetes. Part 4. Investigations on Entorrhiza casparyana by light and electron microscopy. Mycologia 73: 392–398.
Denchev CM, Denchev TT (2012) New records of smut fungi. 6. Mycotaxon 121: 215–223.
Fineran JM (1978) A taxonomic revision of the genus Entorrhiza C. Weber (Ustilaginales). Nova Hedwigia 30: 1–68.
Fineran JM (1980) The structure of galls induced by Entorrhiza C. Weber (Ustilaginales) on roots of the Cyperaceae and Juncaceae. Nova Hedwigia 32: 265–284.
Gargas A, Taylor JW (1992) Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia 84: 589–592.
Garnica S, Riess K, Bauer R, Oberwinkler F, Weiß M (2013) Phylogenetic diversity and structure of sebacinoid fungi associated with plant communities along an altitudinal gradient. FEMS Microbiology Ecology 83: 265–278.
Goker M, Voglmayr H, Blázquez GG, Oberwinkler F (2009) Species delimitation in downy mildews: the case of Hyaloperonospora in the light of nuclear ribosomal ITS and LSU sequences. Mycological Research 113: 308–325.
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511–518.
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780.
Liu M, Hambleton S (2013) Laying the foundation for a taxonomic review of Puccinia coronata s.l. in a phylogenetic context. Mycological Progress 12: 63–89.
Lutz M, Piatek M, Kemler M, Chlebicki A, Oberwinkler F (2008) Anther smuts of Caryophyllaceae: molecular analyses reveal further new species. Mycological Research 112: 1280–1296.
Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis. Version 2.75. https://doi.org/mesquiteproject.org.
McTaggart AR, Doungsaard C, Geering ADW, Aime MC, Shivas RG (2015) A co-evolutionary relationship exists between Endoraecium (Pucciniales) and its Acacia hosts in Australia. Persoonia 35: 50–62.
Piqtek M, Lutz M, Chater AO (2013) Cryptic diversity in the Antherospora vaillantii complex on Muscari species. IMA Fungus 4: 5–19.
Piqtek M, Lutz M, Ronikier A, Kemler M, Świderska-Burek U (2012) Microbotryum heliospermae, a new anther smut fungus parasitic on Heliosperma pusillum in the mountains of the European Alpine System. Fungal Biology 116: 185–195.
Ploch S, Telle S, Choi YJ, Cunnington JH, Priest M, et al. (2011) The molecular phylogeny of the white blister rust genus Pustula reveals a case of underestimated biodiversity with several undescribed species on ornamentals and crop plants. Fungal Biology 115: 214–219.
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313.
Talbot PHB (1956) Entorrhiza calospora sp. nov, and some other parasitic fungi in Limeum roots. Bothalia 6: 453–463.
Vanky K (1994) European Smut Fungi. Jena: Gustav Fischer.
Vanky K (2012) Smut Fungi of the World. St Paul, MN: American Phytopathological Society Press.
Vanky K (2013) Illustrated Genera of Smut Fungi. 3rd edn. St Paul, MN: American Phytopathological Society Press.
Vanky K, Bauer R, Begerow D (2007) Talbotiomyces, a new genus for Entorrhiza calospora (Basidiomycota). Mycologia Balcanica 4: 11–14.
Vasighzadeh A, Zafari D, Selçuk F, Hüseyin E, Kurşat M, et al. (2014) Discovery of Thecaphora schwarzmaniana on Rheum ribes in Iran and Turkey: implications for the diversity and phylogeny of leaf smuts on rhubarbs. Mycological Progress 13: 881–892.
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246.
Voglmayr H, Montes-Borrego M, Landa BB (2014) Disentangling Peronospora on Papaver. phylogenetics, taxonomy, nomenclature and host range of downy mildew of opium poppy (Papaver somniferum) and related species. PLoS ONE 9: e96838.
Weiß M, Sýkorová Z, Garnica S, Riess K, Martos F, et al. (2011) Sebacinales everywhere: previously overlooked ubiquitous fungal endophytes. PLoS ONES: e16793.
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis MA, Gefand DH, Sninsky JJ, White TJ, eds): 315–322. San Diego: Academic Press.
Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: Calibrating the family tree. Proceedings of the Royal Society London B, Biological Sciences 268: 2211–2220.
Zundel GL (1953) The Ustilaginales of the World. [Contribution no.176.] University Park, PN: Pennsylvania State College School of Agriculture.
Acknowledgements
Robert Bauer (1950–2014) initiated this project and unexpectedly passed away before the manuscript was published. We thank Roger G. Shivas (BRIP) for kindly providing a fragment of isotype of Talbotiomyces calosporus from HUV (smut fungi collection of Kálmán Vánky), now deposited in BRIP. We are grateful to Sigisfredo Garnica (Tübingen University), Alistair McTaggart (FABI) and Roger G. Shivas for valuable comments on the manuscript. This study was funded by the German Research Foundation, DFG (BA 75/3-1; BE 2201/7-1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Riess, K., Bauer, R., Kellner, R. et al. Identification of a new order of root-colonising fungi in the Entorrhizomycota: Talbotiomycetales ord. nov. on eudicotyledons. IMA Fungus 6, 129–133 (2015). https://doi.org/10.5598/imafungus.2015.06.01.07
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.5598/imafungus.2015.06.01.07