Abstract
Corynebacterium terpenotabidum Takeuchi et. al 1999 is a member of the genus Corynebacterium, which contains Gram-positive and non-spore forming bacteria with a high G+C content. C. terpenotabidum was isolated from soil based on its ability to degrade squalene and belongs to the aerobic and non-hemolytic Corynebacteria. It displays tolerance to salts (up to 8%) and is related to Corynebacterium variabile involved in cheese ripening. As this is a type strain of Corynebacterium, this project describing the 2.75 Mbp long chromosome with its 2,369 protein-coding and 72 RNA genes will aid the Genomic Encyclopedia of Bacteria and Archaea project.
Similar content being viewed by others
Introduction
Strain Y-11T (= DSM 444721T) is the type strain of the species Corynebacterium terpenotabidum [1]. It was originally isolated from soil, although the exact source has not been published [2,3]. The genus Corynebacterium is comprised of Gram-positive bacteria with a high G+C content. It currently contains over 80 members [4] isolated from diverse backgrounds like human clinical samples [5] and animals [6], but also from soil [7] and ripening cheese [8].
Within this diverse genus, C. terpenotabidum has been proposed to form a subclade together with C. variabile DSM 20132T and C. nuruki S6-4T, demonstrating 97.4% and 95.9% similarity respectively between the 16S rRNA gene sequences. Information on the strain is scarce. It was isolated for its ability to metabolize the linear triterpene squalene and classified as an Arthrobacter species [2,3], but no further information on the strain was supplied. Neither the origin nor the exact isolation procedures were reported. C. terpenotabidum can cleave squalene yielding geranylacetone [2] but also accepts some squalene derivatives [3].
Here we present a summary classification and a set of features for C. terpenotabidum DSM 44721T, together with the description of the genomic sequencing and annotation.
Classification and features
A representative genomic 16S rRNA sequence of C. terpenotabidum DSM 44721T was compared to the Ribosomal Database Project database [9]. C. terpenotabidum shows highest similarity to C. variabile (97.4%).
Figure 1 shows the phylogenetic neighborhood of C. terpenotabidum in a 16S rRNA based tree. Within the genus Corynebacterium, C. terpenotabidum forms a distinct subclade together with C. variabile and C. nuruki.
Phylogenetic tree highlighting the position of C. terpenotabidum relative to type strains of other species within the genus Corynebacterium. Species with at least one publicly available genome sequence (not necessarily the type strain) are highlighted in bold face. The tree is based on sequences aligned by the RDP aligner and utilizes the Jukes-Cantor corrected distance model to construct a distance matrix based on alignment model positions without alignment inserts, using a minimum comparable position of 200. The tree is built with RDP Tree Builder, which utilizes the Weighbor method [10] with an alphabet size of 4 and length size of 1,000. The building of the tree also involves a bootstrapping process repeated 100 times to generate a majority consensus tree [11]. Rhodococcus equi (X80614) was used as an outgroup.
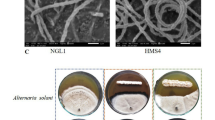
C. terpenotabidum Y-11T cells are Gram-positive non acid fast rods (1.0–1.5 µm × 0.5–0.8 µm wide) that grow strictly aerobically in rough, grayish-white colonies without diffusible pigments or aerial mycelia [1], [Table 1]. Cells grow with a wax-like quality on solid medium and tend to clot in liquid culture. Scanning electron micrograph pictures of liquid grown cultures revealed slight morphological differences between free-floating cells and clotted cells (Figure 2).
C. terpenotabidum was found to be able to utilize fructose, galactose, mannose, lactate, and ethanol as carbon source, while many others like arginine, aspartate, histidine, methylamine, ethylamine, methanol, galactose, lactose, maltose, sucrose, glycerol, sorbitol, mannitol, inositol, citrate, succinate, malonate, pimelate, m-hydroxybenzoate and p-hydroxybenzoate cannot be used. Optimal growth of strain Y-11T is reported at 28°C. C. terpenotabidum was shown to grow with a salinity between 0 and 8.0% (w/v NaCl), with no growth at 10% [1]. The biochemical characterization revealed positive signals for urease, catalase, and hydrolysis of Tween 80.
Chemotaxonomy
The cell wall of C. terpenotabidum Y-11T contains alanine, glutamic acid, and meso-diaminopimelic acid in a molar ratio of 2.12: 1.00: 0.97. The main components of the cell wall sugars are described to be arabinose, galactose, and mannose in a molar ratio of 2.47: 1.71: 1.00. The glycan moiety of the cell wall was found to contain acetyl residues [1].
In C. terpenotabidum, cellular fatty acids are composed mainly of oleic acid (C18:1 ω9c, 31%), palmitic acid (C16:0, 28%), and tuberculostearic acid 10-methyl (C18:0, 21%). The whole-cell methanolysate of strain Y-11 contained mycolic esters [1]. The predominant isoprenoid quinone is menaquinone MK-9(H2).
Genome sequencing and annotation
Genome project history
C. terpenotabidum Y-11T was selected for sequencing as part of a project to define the core genome and pan genome of the non-pathogenic corynebacteria. While not being part of the Genomic Encyclopedia of Bacteria and Archaea (GEBA) project [23], sequencing of the type strain will nonetheless aid the GEBA effort. The genome project is deposited in the Genomes OnLine Database [24] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the Center of Biotechnology (CeBiTec). A summary of the project information is shown in Table 2.
Growth conditions and DNA isolation
C. terpenotabidum strain Y-11T, DSM 44721, was grown aerobically in LB broth (Carl Roth GmbH, Karlsruhe, Germany) at 30 °C. DNA was isolated from ∼ 108 cells using the protocol described by Tauch et al. 1995 [25].
Genome sequencing and assembly
The genome was sequenced using a 454 sequencing platform. A standard 3k paired end sequencing library was prepared according to the manufacturers protocol (Roche). The genome was sequenced using the GS-FLX platform with Titanium chemistry, yielding 384,252 total reads, providing 29.52× coverage of the genome. Pyrosequencing reads were assembled using the Newbler assembler v2.3 (Roche). The initial Newbler assembly consisted of 22 contigs in six scaffolds. Analysis of the six scaffolds revealed five that made up the chromosome, while the remaining one contained five copies of the RRN operon that caused the scaffold breaks. The scaffolds were ordered based on alignments to the complete genomes of C. variabile [26] and subsequent verification by restriction digestion, Southern blotting and hybridization with a 16S rDNA specific probe.
The Phred/Phrap/Consed software package [27–30] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, gaps between contigs were closed by editing in Consed (for repetitive elements) and by PCR with subsequent Sanger sequencing (IIT Biotech GmbH, Bielefeld, Germany). A total of 12 additional reactions were necessary to close gaps not caused by repetitive elements.
To raise the quality of the assembled sequence, Illumina reads were used to correct potential base errors and increase consensus quality. A WGS library was prepared using the Illumina-Compatible Nextera DNA Sample Prep Kit (Epicentre, WI, U.S.A) according to the manufacturer’s protocol. The library was sequenced in a 2x 120 bp paired read run on the MiSeq platform, yielding 2,307,926 total reads. Together, the combination of the Illumina and 454 sequencing platforms provided 91.2× coverage of the genome.
Genome annotation
Gene prediction and annotation were done using the PGAAP pipeline [31]. Genes were identified using GeneMark [32], GLIMMER [33], and Prodigal [34]. For annotation, BLAST searches against the NCBI Protein Clusters Database [35] are performed and the annotation is enriched by searches against the Conserved Domain Database [36] and subsequent assignment of coding sequences to COGs. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [37], Infernal [38], RNAMMer [39], Rfam [40], TMHMM [41], and SignalP [42].
Genome properties
The genome consists of one circular chromosome of 2,751,233 bp (67.02% G+C content) with no additional extrachromosomal elements present. A total of 2,441 genes were predicted, 2,369 of which are protein coding genes. 1,306 (55.13%) of the protein coding genes were assigned to a putative function with the remaining annotated as hypothetical proteins. In addition, 910 protein coding genes belong to 281 paralogous families in this genome, corresponding to a gene content redundancy of 38.41% [Figure 3]. The properties and the statistics of the genome are summarized in Table 3, and Table 4.
References
Takeuchi M, Sakane T, Nihira T, Yamada Y, Imai K. Corynebacterium terpenotabidum sp. nov., a bacterium capable of degrading squalene. Int J Syst Bacteriol 1999; 49:223–229. PubMed http://dx.doi.org/10.1099/00207713-49-1-223
Yamada Y, Motoi H, Kinoshita S, Takada N, Okada H. Oxidative degradation of squalene by Arthrobacter species. Appl Microbiol 1975; 29:400–404. PubMed
Yamada Y, Kusuhara N, Okada H. Oxidation of linear terpenes and squalene variants by Arthrobacter sp. Appl Environ Microbiol 1977; 33:771–776. PubMed
Euzéby JP. List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol 1997; 47:590–592. PubMed http://dx.doi.org/10.1099/00207713-47-2-590
Renaud FNR, Aubel D, Riegel P, Meugnier H, Bollet C. Corynebacterium freneyi sp. nov., alpha-glucosidase-positive strains related to Corynebacterium xerosis. Int J Syst Evol Microbiol 2001; 51:1723–1728. PubMed http://dx.doi.org/10.1099/00207713-51-5-1723
Collins MD, Hoyles L, Foster G, Falsen E. Corynebacterium caspium sp. nov., from a Caspian seal (Phoca caspica). Int J Syst Evol Microbiol 2004; 54:925–928. PubMed http://dx.doi.org/10.1099/ijs.0.02950-0
Zhou Z, Yuan M, Tang R, Chen M, Lin M, Zhang W. Corynebacterium desert sp. nov., isolated from desert sand. Int J Syst Evol Microbiol 2012; 62:791–794. PubMed http://dx.doi.org/10.1099/ijs.0.030429-0
Brennan NM, Brown R, Goodfellow M, Ward AC, Beresford TP, Simpson PJ, Fox PF, Cogan TM. Corynebacterium mooreparkense sp. nov. and Corynebacterium case sp. nov., isolated from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 2001; 51:843–852. PubMed http://dx.doi.org/10.1099/00207713-51-3-843
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37(Database issue):D141–D145. PubMed http://dx.doi.org/10.1093/nar/gkn879
Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol 2000; 17:189–197. PubMed http://dx.doi.org/10.1093/oxfordjournals.molbev.a026231
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 2007; 35(Database issue):D169–D172. PubMed http://dx.doi.org/10.1093/nar/gkl889
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119–169.
Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479–491. http://dx.doi.org/10.1099/00207713-47-2-479
Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 2009; 59:589–608. PubMed http://dx.doi.org/10.1099/ijs.0.65780-0
Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225–420. http://dx.doi.org/10.1099/00207713-30-1-225
Buchanan RE. Studies in the nomenclature and classification of bacteria. II. The primary subdivisions of the Schizomycetes J Bacteriol 1917; 2:155–164. PubMed
Lehmann KB, Neumann R. Lehmann’s Medizin, Handatlanten. X Atlas und Grundriss der Bakteriologie und Lehrbuch der speziellen bakteriologischen Diagnostik., Fourth Edition, Volume 2, J.F. Lehmann, München, 1907, p. 270.
Lehmann KB, Neumann R. Atlas und Grundriss der Bakteriologie und Lehrbuch der speziellen bakteriologischen Diagnostik, First Edition, J.F. Lehmann, München, 1896, p. 1–448.
Bernard KA, Wiebe D, Burdz T, Reimer A, Ng B, Singh C, Schindle S, Pacheco AL. Assignment of Brevibacterium stationis (ZoBell and Upham 1944) Breed 1953 to the genus Corynebacterium, as Corynebacterium stationis comb. nov., and emended description of the genus Corynebacterium to include isolates that can alkalinize citrate. Int J Syst Evol Microbiol 2010; 60:874–879. PubMed http://dx.doi.org/10.1099/ijs.0.012641-0
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056–1060. PubMed http://dx.doi.org/10.1038/nature08656
Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346–D354. PubMed http://dx.doi.org/10.1093/nar/gkp848
Tauch A, Kassing F, Kalinowski J, Pühler A. The Corynebacterium xerosis composite transposon Tn5432 consists of two identical insertion sequences, designated IS1249, flanking the erythromycin resistance gene ermCX. Plasmid 1995; 34:119–131. PubMed http://dx.doi.org/10.1006/plas.1995.9995
Schröder J, Maus I, Trost E, Tauch A. Complete genome sequence of Corynebacterium variabile DSM 44702 isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavor generation. BMC Genomics 2011; 12:545. PubMed http://dx.doi.org/10.1186/1471-2164-12-545
Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195–202. PubMed http://dx.doi.org/10.1101/gr.8.3.195
Gordon D. Viewing and editing assembled sequences using Consed. Curr Protoc Bioinformatics 2003;Chapter 11:Unit11 2.
Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
NCBI. 2010 NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP). http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html.
Borodovsky M, Mills R Besemer J, Lomsadze A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr Protoc Bioinformatics 2003;Chapter 4:Unit4 5.
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 1999; 27:4636–4641. PubMed http://dx.doi.org/10.1093/nar/27.23.4636
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed http://dx.doi.org/10.1186/1471-2105-11-119
Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, et al. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res 2009; 37(Database issue):D216–D223. PubMed http://dx.doi.org/10.1093/nar/gkn734
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 2009; 37(Database issue):D205–D210. PubMed http://dx.doi.org/10.1093/nar/gkn845
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964. PubMed
Eddy SR. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 2002; 3:18. PubMed http://dx.doi.org/10.1186/1471-2105-3-18
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100–3108. PubMed http://dx.doi.org/10.1093/nar/gkm160
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33 (Database Issue):D121–124.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567–580. PubMed http://dx.doi.org/10.1006/jmbi.2000.4315
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783–795. PubMed http://dx.doi.org/10.1016/j.jmb.2004.05.028
Acknowledgements
Christian Rückert acknowledges funding through a grant by the Federal Ministry for Education and Research (0316017) within the BioIndustry2021 initiative.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Rückert, C., Albersmeier, A., Al-Dilaimi, A. et al. Genome sequence of the squalene-degrading bacterium Corynebacterium terpenotabidum type strain Y-11T (= DSM 44721T). Stand in Genomic Sci 9, 505–513 (2014). https://doi.org/10.4056/sigs.4588337
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.4588337