Abstract
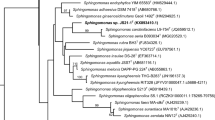
Sphingomonads are becoming increasingly widespread in diverse aquatic and terrestrial environments including the phylosphere and rhizosphere of plants, among which a few have emerged as plant pathogens. The taxonomic status and some biological characteristics of the sphingomonads existing as epiphyte on citrus trees were investigated in the present study. Among the 16 phenogroups, tentatively identified as sphingomonad, nine groups were identified as Sphingomonas, Sphingobium and Novosphingobium strains judged from their 16S rDNA sequences. Four out of the nine groups of strains were identified as Sphingomonas melonis, among which a representative strain was used along with the representatives of the other five groups in MLSA phylogeny employing the housekeeping genes atpD, dnaK, gap, recA, and rpoB. Based on the results of the phylogenies inferred from sequences of concatenated data set, strains of four groups belonged to the genus Sphingomonas, with the ones identified as S. melonis confirmed as such, and the strains of the remaining two groups each were member of Sphingobium and Novosphingobium. The epiphytic existence of S. melonis strains which were able to incite brown spot melon on citrus trees warrants further investigation. All of the strains produced a hypersensitive reaction in geranium and some possessed ice nucleation activity. Further studies are needed to resolve taxonomic status of the unidentified species and to unravel additional biological features of the sphingomonad strains.


Similar content being viewed by others
REFERENCES
Citrus Fruit Fresh and Processed—Statistical Bulletin, Rome: UN Food Agric. Org., 2016.
Ferguson, L. and Grafton-Cardwell, E., Citrus Production Manual, Oakland, CA: Univ. Calif., Agric. Nat. Resour., 2014.
Ashkan, S.M., A Textbook of Fruit Crops Diseases in Iran, Tehran: AIJ, 2006.
Hashem, P., Tajvar, Y., Shekh-Ashkoore, A., Ebadi, H., Fatahi-Moghadam, J., and Faghih-Nasiri, M., Evaluation of Cold and Frost Damage in Citrus and Kiwifruit of Mazandaran Province Citrus, Ramsar: Citrus Subtrop. Fruits Res. Cent., 2017.
Yabuuchi, E., Yano, I., Oyaizu, H., Hashimoto, Y., Ezaki, T., and Yamamoto, H., Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas, Microbiol. Immunol., 1990, vol. 34, no. 2, pp. 99–119.
Takeuchi, M., Hamana, K., and Hiraishi, A., Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses, Int. J. Syst. Evol. Microbiol., 2001, vol. 51, no. 4, pp. 1405–1417.
Jogler, M., Chen, H., Simon, J., Rohde, M., Busse, H.J., Klenk, H.P., Tindall, B.J., and Overmann, J., Description of Sphingorhabdus planktonica gen. nov., sp. nov. and reclassification of three related members of the genus Sphingopyxis in the genus Sphingorhabdus gen. nov, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, no. 4, pp. 1342–1349.
Buonaurio, R., Stravato, V.M., Kosako, Y., Fujiwara, N., Naka, T., Kobayashi, K., Cappelli, C., and Yabuuchi, E., Sphingomonas melonis sp. nov., a novel pathogen that causes brown spots on yellow Spanish melon fruits, Int. J. Syst. Evol. Microbiol., 2002, vol. 52, no. 6, pp. 2081–2087.
Kini, K., Agnimonhan, R., Dossa, R., Soglonou, B., Gbogbo, V., Ouedrago, I., Kpemoua, K., Traore, M., and Silue, D., First report of Sphingomonas sp. causing bacterial leaf blight of rice in Benin, Burkina Faso, the Gambia, Ivory Coast, Mali, Nigeria, Tanzania and Togo, New Dis. Rep., 2017, vol. 35, p. 32.
Kim, H., Nishiyama, M., Kunito, T., Senoo, K., Kawahara, K., Murakami, K and Oyaizu, H., High population of Sphingomonas species on plant surface, J. Appl. Microbiol., 1998, vol. 85, no. 4, pp. 731–736.
Schaad, N.W., Jones, J.B., and Chun, W., Laboratory Guide for Identification of Plant Pathogenic Bacteria, Schaad, N.W., Jones, J.B., and Chun, W., Eds., St. Paul: APS Press, 2001.
Silva, C., Cabral, J.M.S., and Van Keulen, F., Isolation of a beta-carotene over-producing soil bacterium, Sphingomonas sp., Biotechnol. Lett., 2004, vol. 26, no. 3, pp. 257–262.
Rahimian, H., Bacterial leaf spot of zinnia in Mazandaran, J. Sci. Agric., 1995.
Lindow, S.E., Arney, D.C., and Uppr, C.D., Erwinia herbicola: a bacterial ice nucleus active in increasing frost injury to corn, Phytopathology, 1978, vol. 68, no. 3, pp. 523–527.
Verma, H., Rani, P., Kumar Singh, A., Kumar, R., Dwivedi, V., et al., Sphingopyxis flava sp. nov., isolated from a hexachlorocyclohexane (HCH)-contaminated soil, Int. J. Syst. Evol. Microbiol., 2015, vol. 65, no. 10, pp. 3720–3726.
Wilson, K., Preparation of genomic DNA from bacteria, Curr. Protoc. Mol. Biol., 2001, vol. 56, no. 1, pp. 241–245.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J., 16S ribosomal DNA amplification for phylogenetic study, J. Bacteriol., 1991, vol. 173, no. 2, pp. 697–703.
Chen, H., Population structure and species description of aquatic Sphingomonadaceae, PhD Thesis, Munich: Ludwig-Maximilians-University, 2012.
Krishnan, R., Menon, R., Likhitha, R., Busse, H.J., Tanaka, N., Krishnamurthi, S., and Rameshkumar, N., Novosphingobium pokkalii sp nov, a novel rhizosphere-associated bacterium with plant beneficial properties isolated from saline-tolerant pokkali rice, Res. Microbiol., 2017, vol. 168, no. 2, pp. 113–121.
Hall, B.G., Phylogenetic Trees Made Easy: a How-to Manual, Sunderland: Sinauer Associates, 2011.
Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S., MEGA6: Molecular Evolutionary Genetics Analysis version 6.0, Mol. Biol. Evol., 2013, vol. 30, no. 12, pp. 2725–2729.
Nei, M. and Kumar, S., Phylogenetic analysis in molecular evolutionary genetics, Annu. Rev. Genet., 1996, vol. 30, no. 1, pp. 371–403.
Wilantho, A., Deekaew, P., Srisuttiyakorn, C., Tongsima, S., and Somboonna, N., Diversity of bacterial communities on the facial skin of different age-group Thai males, Peer J., 2017, vol. 5, p. e4084.
Kontur, W S., Bingman, C.A., Olmsted, C.N., Wassarman, D.R., Ulbrich, A., Gall, D.L., Smith, R.W., Yusko, L.M., Fox, B.G., Noguera, D.R., Coon, J.J., and Donohue, T.J., Novosphingobium aromaticivorans uses a Nu-class glutathione S-transferase as a glutathione lyase in breaking the β–aryl ether bond of lignin, J. Biol. Chem., 2018, vol. 293, no. 14, pp. 4955–4968.
Liu, F., Zhan, R.L., and He, Z.Q., First report of bacterial dry rot of Mango caused by Sphingomonas sanguinis in China, Plant Dis., 2018, vol. 102, no. 12, p. 2632.
Nikravesh, Z., Arab, F., Rezaeian, V., and Rahimian, H., Isolation of Sphingomonas sp. from peach and willow trees in Kashan, Proc. 17th Iranian Plant Protection Congr., Tehran, 2006, p. 479.
Huang, Y., Feng, H., Lu, H., and Zeng, Y., Novel 16S rDNA primers revealed the diversity and habitats-related community structure of sphingomonads in 10 different niches, Antonie Leeuwenhoek, 2017, vol. 110, no. 7, pp. 877–889.
Pandey, R., Usui, K., Livingstone, R.A., Fischer, S.A., Faendtner, J.P., Backus, E.H.G., Nagata, Y., Fröhlich-Nowoisky, J., Schmüser, L., Mauri, S., Scheel, J.F., Knopf, D.A., Pöschl, U., et al., Ice-nucleating bacteria control the order and dynamics of interfacial water, Sci. Adv., 2016, vol. 2, no. 4, p. e1501630.
Nejad, P., Ramstedt, M., and Granhall, U., Pathogenic ice-nucleating active bacteria in willow for short rotation forestry, For. Pathol., 2004, vol. 34, no. 6, pp. 369–381.
Pallen, M.J. and Wren, B.W., Bacterial pathogenomics, Nature, 2007, vol. 449, no. 7, pp. 835–842.
ACKNOWLEDGMENTS
The authors would like to thank Mrs. Zahra Nikravesh for her excellent assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
About this article
Cite this article
Seyed Zahra Valedsaravi, Rahimian, H., Babaeizad, V. et al. Biological Characteristics and Phylogeny of Sphingomonad Strains Associated with Citrus Trees in Northern Iran. Russ. Agricult. Sci. 47, 606–613 (2021). https://doi.org/10.3103/S1068367421060161
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S1068367421060161