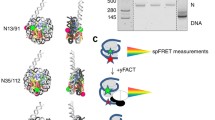
Abstract
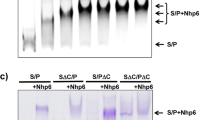
Controlled access to chromatin DNA is required for gene expression and is provided by regulatory factors, such as the FACT protein complex. As was previously shown, in the presence of the Nhp6 protein, yeast FACT (yFACT) performs ATP-independent reversible unfolding of nucleosomes, the mechanism of which requires detailed investigation. In this work, in order to study the mechanism of nucleosome unfolding, it was investigated whether one Nhp6 molecule is sufficient for unwinding nucleosomal DNA by the yFACT factor or whether the reorganization of the nucleosome structure requires the combined action of yFACT and several Nhp6 molecules. The studies carried out by native electrophoresis showed that yFACT can bind at least three Nhp6 molecules. Using single particle microscopy based on Förster resonance energy transfer, it was found that the ability of yFACT to unfold nucleosomes is not increased but decreased with an increase in the yFACT–Nhp6 ratio from 1 : 10 to 1 : 1 at a constant concentration of Nhp6. Therefore, the unfolding of nucleosomes requires the binding of more than one Nhp6 molecule in the nucleosome–yFACT–Nhp6 complex. The data obtained clarify the existing ideas about the reorganization of the nucleosome structure by the yFACT factor.

Similar content being viewed by others
REFERENCES
Clapier, C.R., Iwasa, J., Cairns, B.R., and Peterson, C.L., Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes, Nat. Rev. Mol. Cell Biol., 2017, vol. 18, no. 7, pp. 407–422.
Valieva, M.E., Armeev, G.A., Kudryashova, K.S., Gerasimova, N.S., Shaytan, A.K., Kulaeva, O.I., McCullough, L.L., Formosa, T., Georgiev, P.G., Kirpichnikov, M.P., Studitsky, V.M., and Feofanov, A.V., Large-scale ATP-independent nucleosome unfolding by a histone chaperone, Nat. Struct. Mol. Biol., 2016, vol. 23, no. 12, pp. 1111–1116.
Bondarenko, M.T., Maluchenko, N.V., Valieva, M.E., Gerasimova, N.S., Kulaeva, O.I., Georgiev, P.G., and Studitsky, V.M., Structure and function of histone chaperone FACT, Mol. Biol., 2015, vol. 49, no. 6, pp. 796–809.
Gurova, K., Chang, H.W., Valieva, M.E., Sandlesh, P., and Studitsky, V.M., Structure and function of the histone chaperone FACT—Resolving FACTual issues, Biochim. Biophys. Acta, Gene Regul. Mech., 2018, vol. 1861, no. 9, pp. 892–904.
Chang, H.W., Valieva, M.E., Safina, A., Chereji, R.V., Wang, J., Kulaeva, O.I., Morozov, A.V., Kirpichnikov, M.P., Feofanov, A.V., Gurova, K.V., and Studitsky, V.M., Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins, Sci. Adv., 2018, vol. 4, no. 11, art. ID 2131.
Chang, H.W., Nizovtseva, E.V., Razin, S.V., Formosa, T., Gurova, K.V., and Studitsky, V.M., Histone chaperone FACT and curaxins: effects on genome structure and function, J. Cancer Metastasis Treat., 2019, vol. 5, art. ID 78.
Kantidze, O.L., Luzhin, A.V., Nizovtseva, E.V., Safina, A., Valieva, M.E., Golov, A.K., Velichko, A.K., Lyubitelev, A.V., Feofanov, A.V., Gurova, K.V., Studitsky, V.M., and Razin, S.V., The anti-cancer drugs curaxins target spatial genome organization, Nat. Commun., 2019, vol. 10, no. 1, art. ID 1441.
Orphanides, G., Wu, W.H., Lane, W.S., Hampsey, M., and Reinberg, D., The chromatin-specific transcription elongation factor fact comprises human SPT16 and SSRP1 proteins, Nature, 1999, vol. 400, no. 6741, pp. 284–288.
Thastrom, A., Lowary, P.T., Widlund, H.R., Cao, H., Kubista, M., and Widom, J., Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences, J. Mol. Biol., 1999, vol. 288, no. 2, pp. 213–229.
Gaykalova, D.A., Kulaeva, O.I., Bondarenko, V.A., and Studitsky, V.M., Preparation and analysis of uniquely positioned mononucleosomes, in Chromatin Protocols. Methods in Molecular Biology (Methods and Protocols), Chellappan, S., Ed., New York: Humana Press, 2009, vol. 523, pp. 109–123.
Kudryashova, K.S., Chertkov, O.V., Nikitin, D.V., Pestov, N.A., Kulaeva, O.I., Efremenko, A.V., Solonin, A.S., Kirpichnikov, M.P., Studitsky, V.M., and Feofanov, A.V., Preparation of mononucleosomal templates for analysis of transcription with RNA polymerase using spFRET, in Chromatin Protocols. Methods in Molecular Biology, Chellappan, S., Ed., New York: Humana Press, 2015, vol. 1288, pp. 395–412.
Liu, Y., Zhou, K., Zhang, N., Wei, H., Tan, Y.Z., Zhang, Z., Carragher, B., Potter, C.S., D’Arcy, S., and Luger, K., FACT caught in the act of manipulating the nucleosome, Nature, 2020, vol. 577, no. 7790, pp. 426–431.
Ruone, S., Rhoades, A.R., and Formosa, T., Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes, J. Biol. Chem., 2003, vol. 278, no. 46, pp. 45288–45295.
Funding
This work was financially supported by the Russian Science Foundation (project no. 19-44-02013) and the Department of Science and Technology of India (bilateral project no. GAP0330).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interests. This article does not contain any studies involving humans or animals performed by any of the authors.
Additional information
Translated by G. Levit
About this article
Cite this article
Sivkina, A.L., Feofanov, A.V., Kirpichnikov, M.P. et al. Role of the Nhp6 Protein in Nucleosome Unfolding by the FACT Factor. Moscow Univ. Biol.Sci. Bull. 76, 191–195 (2021). https://doi.org/10.3103/S009639252104012X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S009639252104012X