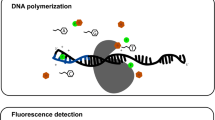
Abstract
On the surface of the nucleosome, there are many regions for interaction with chromatin proteins, but an acidic patch, a negatively charged region formed by the residues of the histone H2A/H2B dimer, is especially prominent. The acidic patch is a target for anticancer drugs and an interaction locus for various pathogens. For instance, it was shown that this region is the binding site of the LANA protein of human gammaherpesvirus 8 and tethers the virus episomes to mitotic chromosomes. The development of methods for analyzing the binding of various compounds to nucleosomes is necessary both for understanding the mechanisms of chromatin regulation and for therapeutic agents' design. In this work, we propose a technique and measure the binding constant of the LANA protein fragment to the nucleosome. In contrast to previous studies, the measurements were carried out at a physiological salt concentration in the buffer solution. The proposed technique is based on the signal detection from the fluorescently labeled peptides after the separation of complexes of nucleosomes with peptides by gel electrophoresis. The paper also discusses mathematical models for analyzing the interaction between peptides and nucleosome and possible factors that can affect it.

Similar content being viewed by others
Change history
18 May 2021
An Erratum to this paper has been published: https://doi.org/10.3103/S0096392521110018
REFERENCES
Horn, V., Recognition of nucleosomes by chromatin factors: Lessons from data-driven docking-based structures of nucleosome-protein complexes, in Chromatin and Epigenetics, Logie, C. and Knoch, T.A., Eds., IntechOpen, 2018. https://doi.org/10.5772/intechopen.81016
Armeev, G.A., Gribkova, A.K., Pospelova, I., Komarova, G.A., and Shaytan, A.K., Linking chromatin composition and structural dynamics at the nucleosome level, Curr. Opin. Struct. Biol., 2019, vol. 56, pp. 46–55.
Kalashnikova, A.A., Porter-Goff, M.E., Muthurajan, U.M., Luger, K., and Hansen, J.C., The role of the nucleosome acidic patch in modulating higher order chromatin structure, J. R. Soc. Interface, 2013, vol. 10, no. 82, 20121022.
Barbera, A.J., Chodaparambil, J.V., Kelley-Clarke, B., Joukov, V., Walter, J.C., Luger, K., and Kaye, K.M., The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA, Science, 2006, vol. 311, no. 5762, pp. 856–861.
Adhireksan, Z., Palermo, G., Riedel, T., Ma, Z., Muhammad, R., Rothlisberger, U., Dyson, P.J., and Da-vey, C.A., Allosteric cross-talk in chromatin can mediate drug-drug synergy: 1, Nat. Commun., 2017, vol. 8, no. 1, 14860.
Beauchemin, C., Moerke, N.J., Faloon, P., and Kaye, K.M., Assay development and high-throughput screening for inhibitors of Kaposi’s sarcoma-associated herpesvirus N-terminal latency-associated nuclear antigen binding to nucleosomes, J. Biomol. Screen, 2014, vol. 19, no. 6, pp. 947–958.
Teles, K., Fernandes, V., Silva, I., Leite, M., Grisolia, C., Lobbia, V.R., van Ingen, H., Honorato, R., Lopes-de-Oliveira, P., Treptow, W., and Santos, G., Nucleosome binding peptide presents laudable biophysical and in vivo effects, Biomed. Pharmacother., 2020, vol. 121, 109678.
Gaykalova, D.A., Kulaeva, O.I., Bondarenko, V.A., and Studitsky, V.M., Preparation and analysis of uniquely positioned mononucleosomes, in Chromatin Protocols. Methods in Molecular Biology (Methods and Protocols), Chellappan, S., Ed., Clifton: Humana Press, 2009, vol 523, pp. 109–123.
Lowary, P.T. and Widom, J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning, J. Mol. Biol., 1998, vol. 276, no. 1, pp. 19–42.
Schindelin, J., Arganda-Carreras, I., Frise, E., et al., Fiji: An open-source platform for biological-image analysis, Nat. Methods, 2012, vol. 9, no. 7, pp. 676–682.
Millman, K.J. and Aivazis, M., Python for scientists and engineers, Comput. Sci. Eng., 2011, vol. 13, no. 2, pp. 9–12.
Weiss, J.N., The Hill equation revisited: Uses and misuses, FASEB J., 1997, vol. 11, no. 11, pp. 835–841.
Chodaparambil, J.V., Barbera, A.J., Lu, X., Kaye, K.M., Hansen, J.C., and Luger, K., A charged and contoured surface on the nucleosome regulates chromatin compaction: 11, Nat. Struct. Mol. Biol., 2007, vol. 14, no. 11, pp. 1105–1107.
Lyubitelev, A.V., Kudryashova, K.S., Mikhaylova, M.S., Malyuchenko, N.V., Chertkov, O.V., Studitsky, V.M., Feofanov, A.V., and Kirpichnikov, M.P., Change in linker DNA conformation upon histone H1.5 binding to nucleosome: Fluorescent microscopy of single complexes, Moscow Univ. Biol. Sci. Bull., 2016, vol. 71, no. 2, pp. 108–113.
Shaytan, A.K., Armeev, G.A., Goncearenco, A., Zhurkin, V.B., Landsman, D., and Panchenko, A.R., Coupling between histone conformations and DNA geometry in nucleosomes on a microsecond timescale: atomistic insights into nucleosome functions, J. Mol. Biol., 2016, vol. 428, no. 1, pp. 221–237.
Funding
This work was financially supported by the Russian Science Foundation (grant no. 19-74-30003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
The study was performed without the use of animals and without involving people as subjects.
CONFLICT OF INTEREST
The authors declare that they do not have any conflict of interests.
ADDITIONAL INFORMATION
Bondarenko ORCID http://orcid.org/0000-0002-5009-0863
Shaytan ORCID http://orcid.org/0000-0003-0312-938X
Additional information
Translated by P. Kuchina
The original online version of this article was revised:
1. Page 252, footnote Deceased is considered invalid.
2. Section ADDITIONAL INFORMATION should read as follows: R. V. Novikov and E. A. Bondarenko contributed equally to this work.
3. Section FUNDING should read as follows: This work was financially supported by the Russian Science Foundation (grant no. 19-74-30003).
About this article
Cite this article
Novikov, R.V., Bondarenko, E.A., Malyuchenko, N.V. et al. Determining the Binding Constant of LANA Protein Fragment with Nucleosome. Moscow Univ. Biol.Sci. Bull. 75, 252–256 (2020). https://doi.org/10.3103/S0096392520040070
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0096392520040070