Abstract
The relict character of the Scots pine populations from the Tatra Mts. (Western Carpathians) was formed on scarcely accessible, ecologically extreme habitats, in areas which were not under human activity. An exhaustive genetic analysis of the Scots pine populations from the refugial locations in the Tatra Mts. (Poland) had not yet been. In this study, we characterize the genetic variation and differentiation of the relict Scots pine populations from the Tatra Mts., to provide information on their genetic resources and the conservation implications. Eight paternally inherited chloroplast microsatellite loci were used to investigate the genetic structure of 4 relict populations, which were compared with the natural populations from the Baltic region (5 populations from Lithuania, Latvia and Estonia) and to a northern range from Russia (5 populations from the Kola Peninsula and Arkhangelsk Oblast). The four relict pine populations were characterized by a lower level of genetic variation when compared to the Baltic and northern populations, which was expressed in lower values of particular genetic parameters: numbers of alleles and haplotypes, haplotype genetic diversity, haplotype differentiation, and mean genetic distance between individuals in the population. Our results revealed a very high and significant genetic differentiation between all the analyzed populations, as well as between the three analyzed regions (PhiPT = 8%). Furthermore, the relict populations within a close geographic location showed higher differentiation (PhiPT = 5%) compared to the northern Russian and Baltic populations that were separated by tens and hundreds of kilometers. The relict populations of P. sylvestris from the Tatra Mts. deserve conservation efforts due to the fact that they are an important component if the species-poor Tatra forests.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Scots pine (Pinus sylvestris L.) is the main component of forest ecosystems in Eurasia, and is a species of great ecological and economic importance. This species of pine is characterized by a wide geographical range and extremely high tolerance to various environmental conditions (Labra et al. 2006; Pravdin 1969; Tóth et al. 2017b). In the northern Tatras (in Poland) there are few populations of P. sylvestris, which are considered as the Holocene relics or even glacial relics (Staszkiewicz 1968, 1993; Jankovska and Pokorny 2008). As has been suggested based on the palynological (Latałowa et al. 2004) and genetic records (Tóth et al. 2017a, b), one of the glacial refugium could exist in the Carpathians and their northern foreland. The relict populations from the Western Carpathians (Tatra Mts. in Poland) possess many distinctive morphological features in terms of their cones, seeds and needles (Zajączkowski 1949; Skrzyszewski 2004). The relict character of the Tatra pine was formed on scarcely accessible, ecologically extreme habitats, which allowed it to survive despite eradicating the human activity in these areas (Łysek 1974). It has a number of morphological traits that classify it as v. carpatica, e.g. smaller and lighter cones, lighter seeds, shorter needles, and a thin light bark (Staszkiewicz 1966). To our knowledge, an exhaustive genetic analysis of the Scots pine populations from the refugial locations in the northern Tatra Mts. has not yet been conducted. This is probably because these populations are usually difficult to access (i.e. they grow in an environment of mountain peat-bogs and rocks). On the other hand, due to the fact that these populations were not subject to anthropopression and were excluded from intensive forest management, they could be an important source of genetic information about the original Scots pine gene pool.
Genetic distinctiveness of the southern Tatra populations from Slovakia was found in the studies of Tóth et al. (2017a). Based on combining paleoenvironmental data, vegetation modeling and genetic analysis it has been shown that isolation in glacial refugia and migrations significantly affect shaping the modern genetic diversity of Scots pine in Europe (Cheddadi et al. 2006). Knowledge about the level of genetic diversity present in relict populations is an extremely valuable basis of the data used for preparing and carrying out protective activities, not only for these small and endangered populations but also for the Scots pine as a species in general This is particularly important in the case of a species that has been intensively cultivated in Europe for over 200 years. As a result, there is scarcely any population of P. sylvestris of a natural or even a primary character, which has not been subjected to anthropopressure. Consequently, research on the genetic variation of natural populations, or those not under forest management, is a rarity (Wojnicka-Półtorak et al. 2017a, b).
Chloroplast microsatellites (cpSSRs), alongside other markers such as nuclear SSRs, RFLPs, mitochondrial markers and allozymes, have for many years been the basic and universal tools used to characterize tree genetic resources (Vendramin et al. 1996; Ribeiro et al. 2002; Navascués et al. 2006), including the Scots pine (Provan et al. 1998; Dzialuk and Burczyk 2002; Wojnicka-Półtorak et al. 2017a).
In this study, we use cpSSR markers in order to characterize the genetic variation of relict Scots pine populations from the north of the Tatra Mts. in the Western Carpathians, and compared these with natural populations from the Baltic region (Lithuania, Latvia and Estonia) and from a northern range in Russia (Kola Peninsula and the Arkhangelsk Oblast). The origins and the level of genetic variation of the natural Scots pine populations in these two regions are poorly documented. These populations, which are characterized by specific features mainly resulting from their complex origin (recolonization after glaciation) and related to specific habitat locations e.g. an extreme northern climate and ecological conditions, forest peat-bogs etc. (Pravdin 1969), could provide valuable baseline information for the conservation and genetic resources of the Scots pine. The P. sylvestris populations from the Baltic region are classified as v. rigensis (v. baltica) (Molotkov and Patlaj 1991; Staszkiewicz 1993), a tree with a straight, soaring stem and with dark red bark.
Our main aim was to characterize the genetic resources of the relict populations of Scots pine in the Western Carpathians and compare them to populations originating from the Baltic region and from northern Russia. To our best knowledge, this is the first report on the relict gene pool of Scots pine from the northern Tatra Mts. (Western Carpathians). We hope to provide relevant information for the conservation and management of present and future gene resources.
Materials and methods
Plant materials
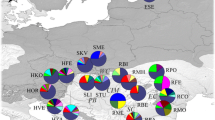
Fourteen populations of Scots pine (420 individuals) covering an area in the northern edge of the range (5 populations), as well as those from the eastern Baltic Sea region (5 populations) were used in the study as the reference populations, and were compared with the relict populations from the northern Tatra Mts. (4 populations from Poland) (Table 1). The Tatra populations of Scots pine were considered as relict and as representing the generations of trees that survived the glaciation (Latałowa et al. 2004; Staszkiewicz 1968, 1993; Jankovska and Pokorny 2008). From each population, needles were collected from 30 randomly selected individuals of Scots pine.
DNA isolation, PCR conditions and capillary electrophoresis
Pine needles frozen in liquid nitrogen (100 mg of tissue) were homogenized and extracted according to the standard CTAB procedures (Doyle and Doyle 1990). The isolated DNA was dissolved in a 0.1 × TE buffer (10 mM pH 8.0 Tris-HCl, 1 mM pH 8.0 EDTA) for further use. The quality and amount of the extracted DNA was checked on a Nanodrop ™ ND-1000 (ThermoScientific) spectrophotometer and diluted to a final concentration of 20 ng / μl. All of the trees were genotyped using eight microsatellite loci, including the following chloroplast SSRs: PCP1289, PCP30277, PCP41131, PCP45071, PCP71987, Pt45002, Pt30204, and Pt71936 (Vendramin et al. 1996; Provan et al. 1998). The characteristics of the analyzed microsatellite loci are presented in Table 2. PCR reactions were performed in a 2720 Thermal Cycler (Applied Biosytems). Two multiplex PCRs were designed. Multiplex I included the following loci: Pt 30204, Pt 71936, PCP 1289, and PCP 41131. In addition, Multiplex II included the loci: PCP 30277, Pt 71987, PCP 45071, and Pt 45002. The PCR conditions were as follows: initial denaturation at 94 °C for 5 min followed by 30 cycles of a 30 s denaturation at 94 °C, annealing 50 °C for 1 min and incubation at 72 °C for 1 min. A final extension was performed at 72 °C for 10 min. The amplification reaction for cpDNA was performed in a 6 μl volume. The reaction mixture contained: 1x Type-it® Microsatellite PCR Kit, 1x Q-Solution, 0.2 μM primers in both directions (F and R), and deionized water. One of the primers of each locus was labeled with a fluorescent marker (6FAM – blue, VIC – green, NED – yellow, and PET – red) to avoid confusion during the analysis of the results. The amplification products were capillary electrophoresed on a 3130xl Genetic Analyzer (Applied Biosystems) using the GeneScan ™ 600LIZ ™ standard. The chromatograms were analyzed in the Peak Scanner ver. 1.0. (Applied Biosystems).
Data analysis
The allelic frequencies, number of alleles and genetic diversity were calculated using FSTAT software (Goudet 2001). Haplotypes were determined as a combination of the different microsatellite variants across the cpSSR loci. Haplotype Analysis ver. 1.05 software (Eliades and Eliades 2009) was used to estimate the haplotype frequencies, number of haplotypes (A), effective number of haplotypes (Ne), haplotypic richness (HR – El Mousadik and Petit 1996), genetic diversity (He), and the mean genetic distance between individuals (Dsh2 – Goldstein et al. 1995). GenAIEx ver. 6.5 software (Peakall and Smouse 2012) was used to: a) divide the total level of genotypic variance into a variance within and among populations and regions by the hierarchical analysis of molecular variance (AMOVA); b) correlate the genetic distance (GD) (Nei 1972) and the geographic distance (GGD) matrices between populations using the Mantel test; and c) calculate pairwise the genetic differentiation between the populations PhiPT (analogue of FST) via AMOVA. The statistical significance of the AMOVA and Mantel tests was determined for all populations and loci, with the number of permutations set to 999.
Results
Allele variation and genetic diversity within populations
A total of 420 samples of P. sylvestris representing fourteen populations were analyzed according to eight chloroplast microsatellite markers, giving a total of 53 size variants, ranging from 3 (PCP41131) to 9 (Pt30204). The distribution of the size variants for each region was unimodal, with the variants differing by 1 bp from each other, which is consistent with a stepwise mutation model (SMM). Those loci were highly polymorphic, except for the locus PCP41131 which was monomorphic in the populations from Lithuania (LZ), Latvia (Ent) and Poland (WK, LS, MG, SKZG). The number of detected alleles per locus was the lowest in the relict populations (mean 3.688), while the northern Russian samples presented the highest values (mean 4.3). The same tendency appeared in the effective number of alleles. The lowest of all values was observed in the Polish relict population (WK, 1.919), while the highest was recorded in the Russian population (RK, 2.995). Private alleles, at a frequency of 0.033, were found in two of the relict populations (WK in locus PT45002; MG in locus Pt71936) and in two Russian populations (RG in locus PCP45071; RG in locus PCP45071) (Table 3).
The average genetic diversity (h) of the eight analyzed cpSSR loci was the lowest (mean 0.480) in the relict populations (from 0.404 to 0.559). In the Baltic and Russian populations, values of 0.533 and 0.562 were found, respectively (Table 3).
The Mantel test showed a significant positive correlation between the genetic and geographic distances for the analyzed populations of Scots pine (R2 = 0.052; P ≤ 0.03).
Haplotype variation
The different size variants resulted in a total of 355 different haplotypes among the 420 individuals. The four relict populations of Scots pine were characterized by the lowest number of haplotypes, with a mean of 27 (ranging from 24 to 30); whereas the populations from the northern edge of the Russian range possessed on average 28 haplotypes (ranging from 27 to 30); and the Baltic populations had 29 haplotypes (ranging from 27 to 30) (see Table 3). The same tendency was reflected in the effective number of haplotypes in the three groups of populations: the highest number was recorded in the Baltic populations (mean 28.4) and the lowest in the relict populations (mean 24.8). A high proportion (50–93%) of the haplotypes found in this study were unique, which means that between 210 to 390 trees (out of 420 individuals) could be uniquely genotyped. The 8% of the haplotypes were detected in two individuals, and the rest were found in 3–8 trees. The frequency of the most common haplotype was the highest in the relict populations (mean 0.075) and lowest in the Baltic populations (mean 0.053). This corresponds to the lowest haplotype differentiation (HH = 0.992) of the relict populations in comparison to the Russian (0.995) and Baltic populations (0.998), as well as the mean genetic distance among individuals (D2sh): 9.27 in the relict; 11.4 in the Baltic; and 13.99 in the Russian populations (see Table 3). There was a different number of haplotypes that was typical for the three groups of populations: in the relict populations it was twelve; and ten in each group of the Russian and Baltic populations. Between the relict and the Russian populations, three common haplotypes were found; between the relict and the Baltic populations there were five; but no common haplotype was found between the Russian and the Baltic populations (see Table 4).
Genetic differentiation among populations
The analysis of molecular variance (AMOVA) locus by locus showed that all of the eight analyzed loci had a significant impact on the level of population differentiation (PhiPT). The highest value of PhiPT was found in two loci: PCP45071 (0.193) and Pt71936 (0.121) (probability 0.01) (Table 2). The AMOVA also revealed high and significant differences among the 14 populations of P. sylvestris, which accounted for 8% (probability of 0.01) of the total cpSSR variance. The variation among the three regions (relict, Baltic and Russian) accounted for about 8% of the total variance and for 3% of the variance among the populations within those groups (probability of 0.01). The AMOVA performed separately within the particular groups showed a very high and significant differentiation among the relict populations (PhiPT = 5%; p < 0.01). The Baltic and Russian populations were much less differentiated: 2% (p < 0.01) and 3% (p < 0.028), respectively (see Table 5).
A PCoA analysis based on the PhiPT values clearly identified three major groups (Fig. 1). One group included the most northern populations from Russia (RM, RO, RK, RG, RW).This group was also distinguished by a high frequency of allele 111 in the locus Pt71987 and of allele 149 in the locus PCP45071. The second group contained the four relict mountain populations from Poland (MG, SKZG, LS, WK) and the third group included the populations from Latvia (LKT, LKm), Lithuania (LZ) and Estonia (Ent, ENm). The first and the second principal coordinates explained 76% of the total variation for the cpSSR markers. A similar pattern of genetic relationships between the Scots pine populations defined above was shown at the dendrogram constructed on the basis of Nei’s (1972) genetic distances (UPGMA method) (Fig. 2). A Principal Coordinate Analysis performed for the relict populations based on the pairwise PhiPT values confirmed their genetic differentiation explaining 98% of the variation (Fig. 3).
Principal coordinate analysis (PCoA) based on the PhiPT values for the 14 analyzed populations (for the abbreviations, see Table 1)
Dendrogram based on Nei’s genetic distances showing the genetic relatedness of the Pinus sylvestris populations (for the abbreviations, see Table 1)
Principal coordinate analysis (PCoA) based on the PhiPT values for the relict populations (for the abbreviations, see Table 1)
Discussion
For the first time, the genetic variation and differentiation of Scots pine populations from the refugial stands in the Tatra Mountains in Poland was assessed based on the polymorphism of cpSSR markers and was compared to the variations found in natural populations from the Baltic region and the northern edge of the species distribution. The comparison with populations representing the Baltic region and the northern extremity of the Scots pine range (from a northern Russian region) makes it possible to assess the variations found in the populations with different histories that evolved under varying selective pressures in different environments. Our data showed that most of the genetic variation located within the Scots pine populations was typical to that of woody species (Hamrick et al. 1992). There were, however, some noticeable differences in the parameters describing the within-population genetic variation of the relict populations vs. the peripheral populations and the populations from the core distribution area.
The four relict pine populations were characterized by a lower level of genetic variation when compared to the Baltic and northern populations, which was expressed in lower values of particular genetic parameters: numbers of alleles and haplotypes, haplotype genetic diversity, haplotype differentiation, and mean genetic distance between individuals in the population. The highest values of these parameters were found in the northern Russian populations. The relict populations had the lowest number of private haplotypes and two private alleles were recorded. By comparison, an excess of private alleles was observed in the peripheral Scots pine populations from Spain and Italy based on an analysis of the allozyme and nSSR loci (Prus-Głowacki et al. 2006; Belletti et al. 2012), as well as in those from Scotland, and the Iberian and Anatolian Peninsulas based on the nSSR loci (Wójkiewicz et al. 2016). Our findings indicated a higher haplotype diversity (HH = 0.984–1.000) and genetic distance between individuals in the populations (D2sh = 6.7–11.16) than was found in the previous studies on Scots pine populations representing a wide distribution range of the Scots pine in Europe and Asia (Semerikov et al. 2014: HH = 0.924–0.992, D2sh = 2.5–6.0; Wójkiewicz and Wachowiak 2016: HH = 0.98–1.00, D2sh = 4.15–11.24). Whereas the long distance gene flow occurring by pollen migration tends to have a homogenizing effect on the genetic variation of Scots pine populations, our results revealed a very high and significant differentiation between all the analyzed populations, as well as between the three analyzed regions (PhiPT = 8%). The relict populations were also significantly differentiated from each other (PhiPT = 5%). In contrast, the populations from the Baltic and northern Russia regions were characterized by lower PhiPT values typical for the cpSSRs (2% and 3% respectively). Our results also showed values considerably higher than those previously reported for P. sylvestris using chloroplast markers (PhiPT = 3.2%, Provan et al. 1998; PhiPT = 0.31%, Robledo-Arnuncio et al. 2005). Previous research on the variation of native populations of P. sylvestris across its broad European distribution using cpSSR markers revealed a much lower inter-population genetic differentiation: from 0.0 to 8% (Wójkiewicz and Wachowiak 2016). Similarly low values of the PhiPT (0.41–1.7%) were found by Semerikov et al. (2014) between 38 Scots pine populations in Asia and Eastern Europe.
The presented findings indicate contrasting trends in genetic differentiation among the populations within various geographical regions. The relict populations with a geographically close location showed higher genetic differentiation compared to the northern Europe and Baltic populations, which were separated by tens and hundreds of kilometers. A similar conclusion was presented by Wójkiewicz et al. (2016) for populations of Scots pine from Europe and Asia Minor. Significant variation between mountain Scots pine populations (14%) was also reported in populations from the Apennines and Alps (Scalfi et al. 2009). In the case of mountain populations from Bulgaria, an AMOVA analysis revealed that a 10.99% variation was found among the population (Naydenov et al. 2005).
The analysis of the PCoA, based on the PhiPT coefficient, pointing out a genetic similarity of the SKZG and MG populations and a separateness of the WK and LS populations. The studies by Staszkiewicz (1966, 1993), based on the morphological variability of cones, proved the existence of two types of P. sylvestris in the Tatra Mts.: slightly changed v. polonica in LS (Łysa Skała) population; and V. meridionalis in the WK (Wielkie Koryciska) population. The separateness of these populations has also been confirmed by the studies of Androsiuk and Urbaniak (2006) based on the needle morphological traits.
Such a large genetic differentiation of populations located at a relatively near geographical distance results from the interaction of many factors, and is known as the “mountain effect” (Naydenov et al. 2011; Schoettle et al. 2012; Celiński et al. 2015) or “mountain-mechanical” (Sannikov et al. 2011). One of these factors is a limitation of the gene flow among isolated mountain localities. The same conclusion was presented by Jadwiszczak et al. (2012) in a study of the glacial relict species Betula nana from the mountain stands in Poland. A restricted gene flow in the relict populations of P. sylvestris seems to be confirmed by the highest frequency of the most common haplotype (mean 0.081) and the lowest mean genetic distance between the individuals in these populations (mean 9.27) compared to the Baltic (CH = 0.053; D2sh = 11.44) and the Russian regions (CH = 0.067; D2sh = 13.99). The adaptation of the Scots pine to low temperatures could have been a crucial factor, allowing the survival of these populations in the mountain refugial area. Lowered values of the genetic diversity parameters in the marginal populations of Betula nana located in the Arctic archipelago of Svalbard were similarly explained as being a consequence of strong climatic selection (Alsos et al. 2002).
The relict populations, even though showing a slight erosion of their chloroplast gene variability, display private alleles and haplotypes and have maintained a sufficient genetic variability to be considered worthy of preservation. The potential role of populations from refugial locations at a higher latitude in the expansion to their present distribution ranges underline the results of studies on cold tolerant species such as Picea abies and Betula pendula (Maliouchenko et al. 2007; Palme et al. 2003; Parducci et al. 2012; Tollefsrud et al. 2008; Willis and van Andel 2004). Glacial refugia generally possess a large part of a species’ allelic richness, and the conservation of these relict populations is therefore crucial for the preservation of a species’ genetic resources (Comps et al. 2001). These populations deserve conservation efforts because they are well adapted to local specific conditions (locally adapted haplotypes), as is demonstrated by their genetic features in the presented results, as well as by the morphological features of the cones (Staszkiewicz 1968, 1993) and differentiation of the needles (Urbaniak et al. 2009). In the species-poor Tatra forests, they are also an important component of the landscape. Further studies of these populations using markers of the nuclear and mitochondrial genome would be needed to verify our results.
References
Alsos IG, Engelskjøn T, Brochmann C (2002) Conservation genetics and population history of Betula nana, Vaccinium uliginosum, and Campanula rotundifolia in the Arctic archipelago of Svalbard. Arct Antarct Alp Res 34(4):408–418
Androsiuk P, Urbaniak L (2006) Differentiation of scots pine (Pinus sylvestris L.) populations in the Tatra Mountains based on needle morphological traits. Biodiversity Resour Conserv 3(4):227–231
Belletti P, Ferrazzini D, Piotti A, Monteleone I, Ducci F (2012) Genetic variation and divergence in Scots pine (Pinus sylvestris L.) within its natural range in Italy. Eur J For Res 131:1127–1138. https://doi.org/10.1007/s10342-011-0584-3
Celiński K, Zbrankova V, Wojnicka-Półtorak A, Chudzińska E (2015) Biogeography and evolutionary factors determine genetic differentiation of Pinus mugo (Turra) in the Tatra Mountains (Central Europe). J Mt Sci 12(3):549–557. https://doi.org/10.1007/s11629-014-3028-y
Cheddadi R, Vendramin GG, Litt T et al (2006) Imprints of glacial refugia in the modern genetic diversity of Pinus sylvestris. Glob Ecol Biogeogr 15:271–282
Comps B, Gömöry D, Letouzey J, Thiébaut B, Petit RJ (2001) Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157:389–397. https://doi.org/10.1126/science.1167936
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dzialuk A, Burczyk J (2002) Comparison of genetic diversity of Scots pine (Pinus sylvestris L.) from qualified seed tree stand and clonal seed orchard. Ecol Quest 2:89–94
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839. https://doi.org/10.1007/BF00221895
Eliades N-G, Eliades DG (2009) Haplotype analysis: software for analysis of haplotype data. Distributed by the authors. In: Forest genetics and Forest tree breeding. Georg-August University Goettingen, Goettingen
Goldstein DB, Linares AR, Cavallisforza LL, Feldman MW (1995) An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471
Goudet J (2001) FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices Version 2.9.3. Available from https://www2.unil.ch/popgen/softwares/fstat.htm
Hamrick JL, Godt MJ, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New For 6:95–124
Jadwiszczak KA, Drzymulska D, Banaszek A, Jadwiszczak P (2012) Population history, genetic variation and conservation status of the endangered birch species Betula nana L. in Poland. Silva Fennica 46(4):465–477
Jankovska V, Pokorny P (2008) Forest vegetation of the last full-glacial period in the Western Carpathians (Slovakia and Czech Republic). Preslia 80:307–324
Labra M, Grassi F, Sgorbati S, Ferrari C (2006) Distribution of genetic variability in southern populations of Scots pine (Pinus sylvestris L.) from the Alps to the Apennines. Flora 201:468–476
Latałowa M, Tobolski K, Nalepka D (2004) Pinus L. subgenus Pinus (subgen. Diploxylon (Koehne) Pilger) – Pine. In: Ralska-Jasiewiczowa M, Latałowa M, Wasylikowa K, Tobolski K, Madeyska E, Wright HE Jr, Turner C (eds) Late glacial and Holocene history of vegetation in Poland based on isopollen maps. Szafer Institute of Botany Polish Academy of Science, Kraków, pp 165–177
Łysek S (1974) Sosna Pinus sylvestris L. In: Myczkowski S (ed) Studia Ośrodka Dokumentacji Fizjograficznej. Rodzime drzewa Tatr, vol 3. Wydawnictwo Polskiej Akademii Nauk, Kraków, pp 87–110 (in Polish)
Maliouchenko O, Palme AE, Buonamici A, Vendramin GG, Lascoux M (2007) Comparative phylogeography and population structure of European Betula species: with particular focus on B. pendula and B. pubescens. J Biogeogr 34:1601–1610. https://doi.org/10.1111/j.1365-2699.2007.01729.x
Molotkov PT, Patlaj IN (1991) Systematic position within the genus and intraspecific taxonomy. In: Giertych M, Csaba M (eds) Genetic of scots pine. Akadémiai Kiadó, Budapest, pp 31–40
Navascués M, Vaxevanidou Z, Gonzáles-Martínez CS, Climent J, Gil L, Emerson CB (2006) Chloroplast microsatellites reveal colonization and metapopulation dynamics in the Canary Island pine. Mol Ecol 15(10):2691–2698. https://doi.org/10.1111/j.1365-294X.2006.02960.x
Naydenov KD, Tremblay FM, Alexandrov A, Fenton NJ (2005) Structure of Pinus sylvestris L. populations in Bulgaria revealed by chloroplast microsatellites and terpenes analysis:provenance tests. Biochem Syst Ecol 33:1226–1245. https://doi.org/10.1016/j.bse.2005.07.011
Naydenov KD, Naydenov MK, Tremblay F (2011) Patterns of genetic diversity that result from bottlenecks in Scots pine and the implications for local genetic conservation and management practices in Bulgaria. New For 42:179–193. https://doi.org/10.1007/s10342-015-0881-3
Nei M (1972) Genetic distances between populations. Am Nat 106:283–292. https://doi.org/10.1086/282771
Palme AE, Su Q, Rautenberg A, Manni A, Lascoux M (2003) Postglacial recolonization and cpDNA variation of silver birch, Betula pendula. Mol Ecol 12:201–212. https://doi.org/10.1046/j.1365-294X.2003.01724.x
Parducci L, Jørgensen T, Tollefsrud MM, Elverland E, Alm T, Fontana SL, Bennett KD, Haile J, Matetovici I, Suyama Y, Edwards ME, Andersen K, Rasmussen M, Boessenkool S, Coissac E, Brochmann C, Taberlet P, Houmark-Nielsen M, Larsen NK, Orlando L, Gilbert MT, Kjær KH, Alsos IG, Willerslev E (2012) Glacial survival of boreal trees in northern Scandinavia. Science 335:1083–1086. https://doi.org/10.1126/science.1216043
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research - un update. Bioinformatics 28(19):2537–2539
Pravdin LF (1969) Scots pine: variation, intraspecific taxonomy and selection. Annarbor Humphrey Science Publishers Ltd, London, pp 1–208
Provan J, Soranzo N, Wilson JN, McNicol WJ, Forrest IG, Cottrell J, Powell W (1998) Gene–pool variation in Caledonian and European Scots pine (Pinus sylvestris L.) revealed by chloroplast simple–sequence repeats. Proc R Soc B 265:1697–1705. https://doi.org/10.1098/rspb.1998.0491
Prus-Głowacki W, Chudzińska E, Wojnicka-Półtorak A, Kozacki L, Fagiewicz K (2006) Effects of heavy metal pollution on genetic variation and cytological disturbances in the Pinus sylvestris L. population. J Appl Genet 47(2):99–108. https://doi.org/10.1007/BF03194607
Ribeiro MM, Mariette S, Vendramin GG, Szmidt AE, Plomin C, Kremer A (2002) Comparison of genetic diversity estimates within and among populations of maritime pine using chloroplast simple-sequence repeat and amplified fragment length polymorphism data. Mol Ecol 11(5):869–877. https://doi.org/10.1046/j.1365-294X.2002.01490.x
Robledo Arnuncio JJ, Collada C, Alia R, Gil L (2005) Genetic structure of montane isolates of Pinus sylvestris L. in a Mediterranean refugial area. Journal of Biogeography 32:595–605. https://doi.org/10.1111/j.1365-2699.2004.01196.x
Sannikov SN, Petrova IV, Schweingruber F, Egorov VE, Parpan VT (2011) Genetic differentiation of Pinus mugo Turra and P. sylvestris L. populations in the Ukrainian Carpathians and the Swiss Alps. Russ J Ecol 42:270–276. https://doi.org/10.1134/S1067413611040151
Scalfi M, Piotti A, Rossi M, Piovani P (2009) Genetic variability of Italian southern Scots pine (Pinus sylvestris L.) populations: the rear edge of the range. Eur J Forest Res 128:377–386. https://doi.org/10.1007/s10342-009-0273-7
Schoettle AW, Goodrich BA, Hipkins V et al (2012) Geographic patterns of genetic variation and population structure in Pinus aristata, rocky mountain bristlecone pine. Can J For Res 42:23–37. https://doi.org/10.1139/X11-152
Semerikov VL, Semerikova SA, Dymshakova OS, Zatsepina KG, Tarakanov VV, Tikhonova IV, Ekart AK, Vidyakin AI, Jamiyansuren S, Rogovtsev RV, Kalchenko LI (2014) Microsatellite loci polymorphism of chloroplast DNA of Scots pine (Pinus sylvestris L.) in Asia and Eastern Europe. Russ J Genet 50(6):577–585
Skrzyszewski J (2004) Charakterystyka morfologiczno-przyrostowa sosny zwyczajnej (Pinus sylvestris L.) w polskiej części Karpat i Sudetów. Zeszyty Naukowe AR Kraków, no. 411 (in Polish)
Staszkiewicz J (1966) Rozmieszczenie „europejskich” gatunków sosny. Wiadomości Botaniczne, t. 10/2: 105-114, Kraków (in Polish)
Staszkiewicz J (1968) Investigations on Pinus sylvestris L., from South-Eastern Europe and from Caucasus and its relation to the pine from other territorie of Europe based on morphological variability of cones. Fragmenta Floristica et Geobotanica Polonica 14(3):259–315. https://doi.org/10.1515/ffp-2017-0013
Staszkiewicz J (1993) Zmienność morfologiczna szpilek, szyszek i nasion. In: Białobok S, Boratyński A, Bugała W (eds.) Biologia sosny zwyczajnej. Sorus Poznań-Kórnik (in Polish)
Tollefsrud MM, Kissling R, Gugerli F, Johnsen Ø, Skrøppa T, Cheddadi R, Vander Knaap WO, Latałowa M, Terhürne-Berson R, Litt T, Geburek T, Brochmann C, Sperisen C (2008) Genetic consequences of glacial survival and postglacial colonization in Norway spruce: combined analysis of mitochondrial DNA and fossil pollen. Mol Ecol 17:4134–4150. https://doi.org/10.1111/j.1365-294X.2008.03893.x
Tóth EG, Vendramin GG, Bagnoli F (2017a) High genetic diversity and distinct origin of recently fragmented Scots pine (Pinus sylvestris L.) populations along the Carpathians and the Pannonian Basin. Tree Genet Genomes 13:47. https://doi.org/10.1007/s11295-017-1137-9
Tóth EGY, Köbölkuti ZA, Pedryc A, Höhn M (2017b) Evolutionary history and phylogeography of Scots pine (Pinus sylvestris L.) in Europe based on molecular markers. J For Res 28:637–651. https://doi.org/10.1007/s11676-017-0393-8
Urbaniak L, Przybyla M, Pisarek W, Chudzińska E, Zielinski R, Polok K (2009) Needle anatomy suggests hybridization between the relict turfosa form of Pinus sylvestris L. from the Gazwa peat bog and typical Scots pine. Acta Soc Bot Pol 78(1):29–36. https://doi.org/10.5586/asbp.2009.005
Vendramin GG, Lelli L, Rossi P, Morgante M (1996) A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol Ecol 5:595–598. https://doi.org/10.1111/j.1365-294X.1996.tb00353.x
Willis KJ, van Andel TH (2004) Trees or no trees? The environments of central and eastern Europe during the last glaciation. Quat Sci Rev 23:2369–2387. https://doi.org/10.1016/j.quascirev.2004.06.002
Wójkiewicz B, Wachowiak W (2016) Substructuring of Scots pine in Europe based on polymorphism at chloroplast microsatellite loci. Flora 220:142–149. https://doi.org/10.1016/j.flora.2016.03.005
Wójkiewicz B, Litkowiec M, Wachowiak W (2016) Contrasting pattern of genetic variation in core and peripheral populations of highly outcrossing and wind pollinated forest tree species. AoB Plants 8. https://doi.org/10.1093/aobpla/plw054
Wojnicka-Półtorak A, Celiński K, Chudzińska E (2017a) Genetic diversity among age classes of a Pinus sylvestris (L.) population from the Białowieża primeval Forest, Poland. Forests 8:227. https://doi.org/10.3390/f8070227
Wojnicka-Półtorak A, Celiński K, Chudzińska E (2017b) Genetic differentiation between generations of Pinus sylvestris natural population: a case study from the last European primeval forest. Aust J Forest Sci 134(3):261–220
Zajączkowski M (1949) Studia nad sosną zwyczajną w Tatrach i Pieninach. Studies on the common pine in the Tatra and Pieniny mountains (Western Carpatians). Prace Rolniczo-Leśne PAU 45 (in Polish)
Acknowledgments
This project was supported by the National Science Centre Poland under Grant No. N N305 0191 40.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and animal rights
This work does not contain any studies involving human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflicts of interest.
Informed consent
Informed consent was obtained from all the individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Urbaniak, L., Wojnicka-Półtorak, A., Celiński, K. et al. Genetic resources of relict populations of Pinus sylvestris (L.) in Western Carpathians assessed by chloroplast microsatellites. Biologia 74, 1077–1086 (2019). https://doi.org/10.2478/s11756-019-00255-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11756-019-00255-6