Abstract
The tumor-node-metastasis (TNM) system fails to accurately predict disease recurrence in a considerable number of patients. Although node-negative (stage II) colon cancer is considered to have an overall good prognosis, the 5-year cancer-specific survival is reported at 81–83% in patients who did not have adjuvant chemotherapy. Thus, reliance on node status alone has led to undertreatment in a subgroup of stage II patients with an unfavorable prognosis. The search for new and better prognosticators in stage II colon cancer has suggested several proposed biomarkers of better prognostication and prediction. However, few such biomarkers have reached widespread clinical utility. For the clinician swimming in the sea of emerging biomarkers, it may be hard to recognize the true floating aid from the surrounding debris in the search for more precise decision-making. Proposed markers include microsatellite instability (MSI) and KRAS and BRAF mutations, but a number of gene panels and consensus molecular subtypes are proposed for clinical prediction and prognostication as well. Although several studies suggest such biomarkers or panels to have a prognostic role in subgroups of patients, a number of studies are reported in heterogeneous groups with in part discordant findings, which again distorts the predictive and prognostic ability of each marker. Lack of homogeneous cohorts, underpowered studies in strict subgroups and challenges in analytical and clinical validity may hamper the progress toward widespread clinical utility. The harvest of prognostic biomarkers in colon cancer has yielded a huge number of candidates for which it is now time to separate the wheat from the chaff.
Similar content being viewed by others
Introduction
Although node-negative (stage II) colon cancer is considered to have an overall good prognosis, the 5-year cancer-specific survival is reported at 81–83% in patients who did not have subsequent adjuvant chemotherapy (1), suggesting the presence of risk for recurrence in a subgroup of patients. Currently, stage II high-risk patients are identified through suggested “high-risk” features in consensus guidelines from the American Society for Clinical Oncology, the European Society for Medical Oncology and the National Comprehensive Cancer Network (NCCN) for which pT4 tumors, poor histology differentiation, presentation of perforation (or obstruction) and inadequate lymph node sample (e.g., <12 nodes) for evaluation are shared risk factors in the guidelines (2–4). Notably, the NCCN guidelines exclude patients with poor histology differentiation if they have microsatellite instability (MSI) from the risk group. Also, these guidelines add a “close or indeterminate resection margin” as a criterion for adjuvant chemotherapy. Evident from the guidelines is, on the one side, the lack of uniform criteria, and thus the risk of introducing treatment variation dependent on which guidelines doctors adhere to; on the other side is the emergence of genetic information (e.g., MSI status) to further refine risk groups for adjuvant chemotherapy. Still, considerable under- and overtreatment is expected through the lack of precise prognostic markers.
Although still being firmly debated (5), the lymph node status is still the strongest predictor and prognosticator in the tumor-node-metastasis (TNM) staging system (6). However, although node status has guided decisions on whether to give adjuvant chemotherapy, it fails (alone) to accurately predict disease recurrence in a considerate number of patients and has led to undertreatment in a subgroup of stage II patients with an unfavorable prognosis. Following this line of emerging evidence, the search for new and better prognosticators in stage II colon cancer is ongoing, with several proposed strategies and approaches to find better biomarkers for prognostication and prediction (7–9). For the clinician drowning in the sea of emerging biomarkers, it may be hard to swim when the debris to grasp for does not show whether you will sink or float in the search for more precise decision-making.
Notably, there is a link between the node status and the genetic makeup of a tumor (10,11) that has been linked to prognosis, but with causality yet to be established. In a recent study, in this journal (12), the role of MSI, KRAS, BRAF and PIK3CA in stage II colon cancers was investigated. In the study by Vogelaar et al. (12), MSI was found in 23% of patients with a trend toward reduced disease-free and overall survival for patients with MSI, which is in contrast to the prognostic role of MSI found by others in early-stage colon cancers (13). This contradictory finding obviously warrants explanation based on the background of existing literature from the past couple of decades. Vogelaar et al. also confirmed a trend toward poorer prognosis in patients with mutant KRAS or BRAF (12). The negative impact of these mutations has been noted by others in both early- and late-stage colorectal cancer (CRC) (11,14–16). Strikingly, the high mutation rate of BRAF in the study by Vogelaar et al. (19%) should be noted, as this rate is usually reported as a single-digit percentage, typically at 5–8% (17). However, an unusually high rate of BRAF mutations (15%) has been found by others as well (18). Finally, the results pointed toward trends but lacked statistical significance and included the chance for underpowered samples in subgroups and type II errors in interpretation.
Why is it important to define subgroups of stage II patients? For one, the overall risk for recurrence is not huge but is present in about 20% of stage II patients (19). A small but significant risk reduction is noted by giving adjuvant chemotherapy to stage II patients, but risk for overtreatment is still high, and the currently proposed high-risk criteria are inaccurate (20). Much energy has been put into gene panels that would yield better prognostic and predictive tools for risk assessment (21–23), but the results are thus far disappointing, with no panels having reached widespread incorporation in clinical practice. The same goes for most other proposed prognostic systems (9,19,24–26).
Several explanations may be brought forward as to why available gene panels have not yet reached clinical utility (27). The clinical samples investigated may not have been representative, derived from cohorts of previous diagnostic and therapeutic eras or from randomized trials, where patients’ (supra)-selection entry criteria do not represent the general patient cohorts; or, investigated cohorts have been subject to case mix of both treatment-naive and adjuvant chemotherapy-treated patients. Further, assay development and technology may not have been robust and valid, thus producing variation in findings across studies and hampering validation. Also, beyond analytical and clinical validity is the current lack of demonstration of clinical utility for many proposed markers or panels (28). Indeed, one really has to demonstrate that the prognostic tools work in real life and make a difference for patients in terms of selecting the accurate therapy that works for the particular patient.
Developing new and pragmatic approaches to biomarker research and development to ensure validity and utility relies on standardization of procedures. This is easier said than done: standardizing a complex and long process such as biomarker validation could prove a hefty swim in the sea of variables that arise when working with biological material (29). If not properly accounted for, anything from patients’ inherent physiology and genetic composition to the actual disease state, the handling techniques of the laboratory or the chosen data analysis methods can potentially affect outcomes. As a starting point in standardization, appropriate cohort sub-grouping is of fundamental importance: if done inaccurately, it can dilute the power of studies at best, or at worst, completely invalidate findings.
To recognize emerging subtype consensus classifications and probe their clinical validity as well as prognostic and predictive ability, the TNM system is thus necessary. The immunoscore (30), mentioned by Vogelaar et al. in the discussion of their work, is a tumor classification system based on the analysis of tumor-infiltrating lymphocyte subpopulations. This system has proven promising at predicting metastatic behavior (and thus survival) in a variety of cohorts and, even more interestingly, may not only be prognostic but also highly suggestive of patients amenable to immune-based cancer therapies (31); however, it lacks validation in prospective studies. A task force group has been launched, but validation of data across several cohorts is still awaited (32).
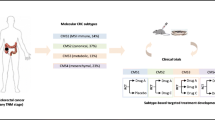
Subdividing patients according to a gene expression-based system is perhaps yet another valuable option. Gene expression data pooled from various studies recently produced a consensus molecular classification of CRC, subdividing it into four classes (33). These clustered around molecular features, such as MSI, hypermutation and immune infiltration (CMS1), chromosomal instability (CMS2-canonical), KRAS mutations and metabolic deregulation (CMS3-metabolic) and a mesenchymal geno- and phenotype (CMS4). Disturbingly, a subcohort of about 7% was not classifiable (labeled as “mixed or indeterminate”) in the proposed set, and a further 7% was classified as mixed groups between the four suggested “pure” groups—thus, further refinement is needed of this proposed classification. In truth, much is still to be learned about the complex genetic and epigenetic makeup of colorectal cancers (34).
Conclusion
Standardized prognostic biomarkers that can reach widespread clinical utility are now needed. Molecular traits are already at the foundations of companion biomarkers research and are increasingly employed to predict which group of patients will respond to any given treatment. Further refinement is needed to avoid under- and overtreatment, prevent patients from harm and ensure effective health economics with increasingly available high-cost drugs. Fast and reliable molecular techniques are now available with increased analytical validity. Clinical validity should be ensured through investigation of correctly powered and well-sized cohorts of homogeneous subgroups in a prospective fashion. Only then will results eventually allow prognostic molecular biomarkers to find a place and be integrated as tumor classification systems beyond the TNM system. We have harvested a huge number of bio-markers for which the prognostic yield has not reach its potential—it is now time to separate the wheat from the chaff for clinical utilization.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Bockelman C, Engelmann BE, Kaprio T, Hansen TF, Glimelius B. (2015) Risk of recurrence in patients with colon cancer stage II and III: A systematic review and meta-analysis of recent literature. Acta Oncol. 54:5–16.
Labianca R, et al. (2013) Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 24(Suppl. 6):vi64–vi72.
Benson AB III, et al. (2004) American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J. Clin. Oncol. 22:3408–19.
Network NCC. (2015) NCCN Guidelines: Colon Cancer, Version 2. https://www.nccn.org/patients/guidelines/colon. National Comprehensive Cancer Network (NCCN); Fort Washington, PA, USA. Accessed August 9, 2016.
Veen T, et al. (2013) Qualitative and quantitative issues of lymph nodes as prognostic factor in colon cancer. Dig. Surg. 30:1–11.
Lea D, Haland S, Hagland HR, Soreide K. (2014) Accuracy of TNM staging in colorectal cancer: A review of current culprits, the modern role of morphology and stepping-stones for improvements in the molecular era. Scand. J. Gastroenterol. 49:1153–63.
Hansen TF, et al. Redefining high-risk patients with stage II colon cancer by risk index and microRNA-21: Results from a population-based cohort. Br. J. Cancer. 111:1285–92.
Fang SH, Efron JE, Berho ME, Wexner SD. (2014) Dilemma of stage II colon cancer and decision making for adjuvant chemotherapy. J. Am. Coll. Surg. 219:1056–69.
Dalerba P, et al. (2016) CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N. Engl. J. Med. 374:211–22.
Berg M, et al. (2015) Molecular subtypes in stage II–III colon cancer defined by genomic instability: Early recurrence-risk associated with a high copy-number variation and loss of RUNX3 and CDKN2A. PLoS One. 10:e0122391.
Berg M, et al. (2013) Influence of microsatellite instability and KRAS and BRAF mutations on lymph node harvest in stage I–III colon cancers. Mol. Med. 19:286–93.
Vogelaar F, et al. (2015) The prognostic value of microsatellite instability, KRAS, BRAF and PIK3CA mutations in stage II colon cancer patients. Mol. Med. 2015:1–26.
Merok MA, et al. (2013) Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: Results from a large, consecutive Norwegian series. Ann. Oncol. 24:1274–82.
Birgisson H, et al. (2015) Microsatellite instability and mutations in BRAF and KRAS are significant predictors of disseminated disease in colon cancer. BMC Cancer. 15:125.
de Cuba EM, et al. (2016) Prognostic value of BRAF and KRAS mutation status in stage II and III microsatellite instable colon cancers. Int. J. Cancer. 138:1139–45.
Soreide K, Sandvik OM, Soreide JA. (2014) KRAS mutation in patients undergoing hepatic resection for colorectal liver metastasis: A biomarker of cancer biology or a byproduct of patient selection? Cancer. 120:3862–65.
Hamilton SR. (2013) BRAF mutation and microsatellite instability status in colonic and rectal carcinoma: Context really does matter. J. Nat. Cancer Inst. 105:1075–77.
Lochhead P, et al. (2013) Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J. Nat. Cancer Inst. 105:1151–56.
Boland CR, Goel A. (2016) Prognostic subgroups among patients with stage II colon cancer. N. Engl. J. Med. 374:277–8.
Dienstmann R, Salazar R, Tabernero J. (2015) Personalizing colon cancer adjuvant therapy: Selecting optimal treatments for individual patients. J. Clin. Oncol. 33:1787–96.
Agesen TH, et al. (2012) ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 61:1560–67.
Yothers G, Song N, George TJ Jr. (2016) Cancer hallmark-based gene sets and personalized medicine for patients with stage II colon cancer. JAMA Oncol. 2:23–24.
Brenner B, et al. (2016) Impact of the 12-gene colon cancer assay on clinical decision making for adjuvant therapy in stage II colon cancer patients. Value Health. 19:82–87.
Jeong DH, Kim WR, Min BS, Kim YW, Song MK, Kim NK. (2015) Validation of a quantitative 12-multigene expression assay (Oncotype DX((R)) Colon Cancer Assay) in Korean patients with stage II colon cancer: implication of ethnic differences contributing to differences in gene expression. Onco. Targets Ther. 8:3817–25.
Cserni G, Bori R, Sejben I, Agoston EI, Acs B, Szasz AM. The Petersen prognostic index revisited in Dukes B colon cancer: Inter-institutional differences. Pathol. Res. Pract. 2016;212(2):73–76.
Zhu J, Dong H, Zhang Q, Zhang S. (2015) Combined assays for serum carcinoembryonic antigen and microRNA-17-3p offer improved diagnostic potential for stage I/II colon cancer. Mol. Clin. Oncol. 3:1315–18.
Bartley AN, Hamilton SR. (2015) Select biomarkers for tumors of the gastrointestinal tract: Present and future. Arch. Pathol. Lab. Med. 139:457–68.
Parkinson DR, et al. (2014) Evidence of clinical utility: An unmet need in molecular diagnostics for patients with cancer. Clin. Cancer Res. 20:1428–44.
de Gramont A, et al. (2015) Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat. Rev. Clin. Oncol. 12:197–212.
Galon J, et al. (2014) Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 232:199–209.
Ascierto PA, et al. (2013) The additional facet of immunoscore: Immunoprofiling as a possible predictive tool for cancer treatment. J. Transl. Med. 11:54.
Galon J, et al. (2012) Cancer classification using the Immunoscore: A worldwide task force. J. Transl. Med. 10:205.
Guinney J, et al. (2015) The consensus molecular subtypes of colorectal cancer. Nat. Med. 21:1350–56.
Okugawa Y, Grady WM, Goel A. (2015) Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 149:1204–25.e12.
Acknowledgments
The work is in part supported from Folke Hermansen Cancer Fund, the Stavanger University Fund and Mjaaland Cancer Fund. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this license, visit (http://creativecommons.org/licenses/by-nc-nd/4.0/)
About this article
Cite this article
Watson, M.M., Søreide, K. The Prognostic Yield of Biomarkers Harvested in Chemotherapy-Naive stage II Colon Cancer: Can We Separate the Wheat from the Chaff?. Mol Med 22, 271–273 (2016). https://doi.org/10.2119/molmed.2016.00098
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2016.00098