Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is an important pathogen in pneumonia associated with severe lung damage. Tissue injury causes release of damage-associated molecular patterns (DAMPs), which may perpetuate inflammation. DNA has been implicated as a DAMP that activates inflammation through Toll-like receptor 9 (TLR9). The aim of this study was to evaluate the role of TLR9 in MRSA pneumonia. Wild-type (Wt) and TLR9 knockout (tlr9−/−) mice were infected intranasally with MRSA USA300 (BK 11540) (5E7 CFU) and euthanized at 6, 24, 48 or 72 h for analyses. MRSA pneumonia was associated with profound release of cell-free host DNA in the airways, as reflected by increases in nucleosome and DNA levels in bronchoalveolar lavage fluid (BALF), accompanied by transient detection of pathogen DNA in MRSA-free BALF supernatants. In BALF, as compared with Wt mice, tlr9−/− mice showed reduced tumor necrosis factor α and IL-6 levels at 6 h and reduced bacterial clearance at 6 and 24 h postinfection. Furthermore, tlr9−/− mice exhibited a greater influx of neutrophils in BALF and increased lung consolidation at 24 and 48 h. This study demonstrates the release of host- and pathogen-derived TLR9 ligands (DNA) into the alveolar space after infection with MRSA via the airways and suggests that TLR9 has proinflammatory effects during MRSA pneumonia associated with enhanced bacterial clearance and limitation of lung consolidation.
Similar content being viewed by others
Introduction
Infectious diseases remain a threat to public health in both developed countries and poor-resource settings. The aging, infection-susceptible population combined with the rise of antibiotic resistant bacteria are important factors herein (1). A striking example of a bacterial threat is Staphylococcus (S.) aureus (2). This gram-positive pathogen is a leading cause of infection worldwide (3) and a major causative pathogen in nosocomial pneumonia (4,5). Roughly half of the world population carries S. aureus on their skin and in the United States alone more than 89 million people are colonized, of whom 2.3 million carry methicillin-resistant S. aureus (MRSA) (6,7). Increasing our understanding of the pathophysiology of MRSA infections is a prerequisite for innovative treatments for this clinically highly relevant microorganism.
MRSA possesses a repertoire of toxins that cause severe pneumonia with massive lung damage that can proceed to a characteristic necrotizing infection (8). During organ damage and cellular necrosis, intracellular structures are released outside the cell, which then can act as damage-associated molecular pattern (DAMP). DAMPs activate pattern recognition receptors (PRRs) and contribute to inflammation as a secondary signal (9,10). Recently, extracellular host DNA has gained increased attention as an important DAMP involved in the pathogenesis of multiple diseases. Systemic inflammation and sepsis result in a dramatic increase in circulating extracellular host DNA (11–13). Immune cells can detect DNA by several sensors in the cytoplasm (14,15). One of these sensors is Toll-like receptor 9 (TLR9), which upon recognition of DNA triggers activation of cells (14,16). TLR9 can also detect bacterial DNA (17–19), including that of MRSA (20). Considering the central role of TLR9 in the recognition of extracellular DNA and the injurious host response associated with cellular damage and death during MRSA pneumonia, we here sought to determine the role of TLR9 in experimentally induced MRSA pneumonia.
Materials and Methods
Mice
All experiments were carried out in accordance with the Dutch Experiment on Animals Act and approved by the Animal Care and Use Committee of the University of Amsterdam (Permit number: DIX102291). Pathogen-free 8-10-wk-old wild-type (Wt) C57BL/6 mice were purchased from Charles River Laboratories Inc. TLR9-deficient (tlr9−/−) mice, backcrossed six times to a C57BL/6 background, were generated as described previously (19). Age- and sex-matched animals were used in all experiments.
Experimental Infection and Tissue Preparation
Pneumonia was induced as described (21,22), using the MRSA strain BK11540 (USA300), kindly provided by Timothy J Foster, Department of Microbiology, Trinity College, Dublin, Ireland. Characteristics of this strain are: multilocus sequence type ST8, spa type 1, staphylococcal cassette chromosome mecIV, agr type 1 and Panton-Valentine leukocidin negative (23). In brief, mice were intranasally inoculated with 107 colony-forming units (CFU)/50 µL saline solution (n = 8 per strain at each time point) under isoflurane (Abbot Laboratories) inhalation anesthesia. The infectious dose is sub-lethal and based on earlier dose-finding studies (21). Mice were killed 6, 24, 48 or 72 h after infection by cardiac puncture under Domitor (Pfizer Animal Health Care, active ingredient medetomidine) and Nimatek (Eurovet Animal Health, active ingredient ketamine) anesthesia. Blood was drawn into EDTA-coated tubes. Bronchoalveolar lavage fluid (BALF) was obtained by exposing the trachea via a midline incision. The trachea was then cannulated with a sterile 22-gauge Abbocath-T catheter (Abbott Laboratories). Unilateral BAL was performed on the left lung by binding the right bronchus and instilling and retrieving 0.7 mL (in aliquots) of sterile phosphate-buffered saline. The right lung was harvested for histology. To obtain lung homogenates, left lungs were excised and homogenized in four volumes of isotonic saline (milliliters) using a tissue homogenizer (Biospec Products). Organs were removed aseptically and homogenized in four volumes of sterile isotonic saline using a tissue homogenizer (Biospec Products). To determine bacterial loads, 10-fold dilutions were plated on blood agar plates and incubated at 37°C for 16 h, after which colonies were counted.
Cell-Free DNA extraction from BALF
After spinning the BALF at 500g and 4°C, for 15 min, the top 500 µL was transferred into a fresh Eppendorf tube, keeping well above the cell pellet while pipetting. Using the harvested BALF, the above procedure was repeated, this time spinning at 20,000g and transferring the top 300 µL of BALF. Cell-free DNA was extracted from this twice-spun BALF using the QIAamp DNA Blood Mini Kit (Qiagen) according to the “Blood and Body Fluid Spin Protocol” recommended by the manufacturer. After binding to the silica matrix and several washing steps, the DNA was eluted with 60 µL of H2O.
MRSA and Mouse Standard Curves
To quantify the amounts of cell-free mouse and MRSA DNA in BALF, standard curves were generated. First, DNA was extracted from both mice toes and MRSA strain BK11540 using the method described above. Next, the amount of DNA in both stocks was measured using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Then, these DNA stocks were used to make eight-step standard curves, starting with 500 ng DNA diluted 10-fold each step. Finally, these standard curves were run together with the samples.
Polymerase Chain Reaction Assays
Oligonucleotides was an Invitrogen product. Primer sequences were as follows: mSOCS-3 forward, ACCTTTC TTATCCGCGACAG; mSOCS-3 reverse, TGCACCAGCTTGAGTACACAG; MRSA forward, TCAACATCTTTC GCATGATTCAACAC; MRSA reverse, CTAGCTTTATTTcAGCAGGTGACGAT. Real-time polymerase chain reaction (RT-PCR) analysis was performed with the StepOnePlus RT-PCR system (Applied Biosystems) using SYBR Green chemistry. The single plex reactions were set up in a volume of 25 µL. DNA primers used at final concentrations of 300 nmol/L. Cycling conditions were 2 min at 50°C and 10 min at 95°C, followed by 50 cycles of denaturation for 15 s at 95°C and primer annealing and elongation for 1 min at 60°C.
Determination of BALF Cell Counts
Cell counts were determined for each BALF sample in a hemocytometer (Beckman Coulter), and differential cell counts were performed on cytospin preparations stained with Giemsa stain (Diff-Quick).
Assays
Nucleosome levels were measured using ELISA as described (24,25). Tumor necrosis factor (TNF-α), interleukin (IL)-1β, IL-6, keratinocyte-derived chemokine (KC) and macrophage inflammatory protein-2 (MIP-2) (all R&D Systems) were measured using specific ELISAs according to manufacturer’s recommendations.
Histology
The left lung lobe was harvested, fixed in 10% buffered formalin and embedded in paraffin. About 4 µm sections were stained with hematoxylin and eosin (HE) and analyzed by a pathologist blinded for groups as described (21,22). To score lung inflammation and damage, the entire lung surface was analyzed with respect to the following parameters: bronchitis, edema, interstitial inflammation, intra-alveolar inflammation, pleuritis, endothelialitis and percentage of the lung surface demonstrating lung consolidation. Each parameter was graded 0–4, with 0 being “absent” and 4 being “severe.” The total pathology score for lungs was expressed as the sum of the score for all parameters.
Statistical Analysis
Data are expressed as box-and-whisker diagrams depicting the smallest observation, lower quartile, median, upper quartile and largest observation unless indicated otherwise. Differences between tlr9−/− and Wt mice were analyzed by Mann-Whitney U test. Analyses were done using GraphPad Prism version 5.0, GraphPad Software. Values of p < 0.05 were considered as statistically significant difference.
All supplementary materials are available online at www.molmed.org .
Results
Host- and Pathogen-Derived DNA Is Released in the Airways During MRSA Pneumonia
To examine whether MRSA pneumonia is associated with the local release of cell-free host DNA, we measured nucleosomes in BALF obtained before and up to 72 h after infection (Figure 1A). BALF of uninfected mice contained very low nucleosome concentrations. After infection with MRSA via the airways, nucleosome levels increased more than 10-fold in BALF within 6 h and kept rising during the course of the infection, reaching peak levels 72 h postinfection that were approximately 1,000-fold higher than in uninfected mice (p < 0.001). As an alternative way to measure cell-free host DNA, we performed a PCR on mouse suppressor of cytokine signaling-3 (SOCS-3) in BALF obtained 6, 24 and 48 h after infection (Figure 1B). Host DNA was detectable after 6 h and, similar to the nucleosome measurements, increased nearly 10-fold after 24 and 48 h. MRSA pneumonia was also associated with elevated levels of MRSA DNA in BALF (Figure 1C); the highest levels were found early after infection (6 h), decreasing to undetectable levels at 48 h postinfection. Together, these data show that MRSA pneumonia is associated with release of both pathogen- and host-derived DNA into BALF, whereby host DNA in particular appears late during infection, while pathogen DNA is found especially early after infection.
Nucleosome concentrations in mouse BALF during S. aureus pneumonia. Wt mice were intranasally infected with 1 × 107 CFU S. aureus and euthanized at the indicated time points. (A) Nucleosome concentrations in BALF quantified by ELISA and expressed as arbitrary units. (B) Host cell-free DNA (mouse SOCS-3) in BALF quantified by PCR. (C) S. aureus DNA in cell-free BALF quantified by PCR. Data represent the means ± standard error of the mean (n = 6–8 mice per time point). *p < 0.05, **p < 0.01 and ***p < 0.001 versus naïve mice (0 h).
TLR9 Facilitates Bacterial Clearance from the Lungs after Infection with S. aureus
To determine the role of TLR9 in clearance of MRSA after induction of pneumonia, we quantified bacterial loads in lungs, BALF, liver and blood of tlr9−/− and Wt mice after intranasal inoculation with MRSA. Tlr9−/− mice showed up to 10 times higher bacterial loads compared with Wt mice at 6 and 24 h after infection in lung homogenates (Figure 2A, p < 0.01 and p < 0.001, respectively) and BALF (Figure 2B, p < 0.01 for both time points). Liver bacterial loads were similar in tlr9−/− and Wt mice at 6 and 24 h postinfection and undetectable later on (Figure 2C); blood remained sterile in blood of both mouse strains at all time points.
Bacterial clearance in tlr−/− mice is reduced after infection with S. aureus via the airways. Bacterial loads after intranasal infection with 1 × 107 CFU S. aureus in (A) lung and (B) BALF of Wt mice (gray) and tlr9−/− mice (white). Data are expressed as box-and-whisker diagrams depicting the median, the smallest observation, lower quartile, median, upper quartile and largest observation (n = 8 mice per group at each time point). **p < 0.01 and ***p < 0.001 versus Wt mice at the same time point.
TLR9 Contributes to the Early Pulmonary Cytokine Response after Infection with MRSA
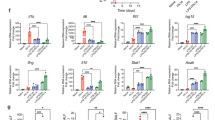
To investigate a role for TLR9 in the inflammatory response to MRSA in the airways, we measured the proinflammatory cytokines TNF-α, IL-1β and IL-6 in BALF from tlr9−/− mice and Wt mice at multiple time points after the induction of pneumonia (Figure 3). All cytokines reached peak concentrations at 6 h after infection. Consistent with a role for TLR9 in the induction of innate immunity during S. aureus pneumonia, and in spite of higher bacterial loads, tlr9−/− mice showed decreased TNF-α (Figure 3A) and IL-6 concentrations (Figure 3B) in BALF versus Wt mice (p < 0.01 and p < 0.05, respectively) at 6 h postinfection, whereas IL-1β concentrations were similar in both groups (Figure 3C). Remarkably, whereas TNF-α was undetectable in BALF at later time points, BALF IL-6 and IL-1β levels were in higher in tlr9−/− mice at 24 h after infection versus Wt mice (both p < 0.001). At later time points, IL-6 and IL-1β concentrations were similar in both mouse strains.
The early cytokine response to S. aureus is reduced in the airways of tlr9−/− mice. (A-C) Cytokine (TNF-α, IL-6 and IL-1 β) levels in BALF at different time points after intranasal infection of 1 × 107 CFU S. aureus in Wt mice (gray) and tlr9−/− mice (white). Data are expressed as box-and-whisker diagrams depicting the median, the smallest observation, lower quartile, median, upper quartile and largest observation (n = 7–8 mice per group at each time point). *p < 0.05, **p < 0.01 versus Wt mice at the same time point.
TLR9 Deficiency Results in Increased Neutrophils Numbers in the Alveolar Space During MRSA Pneumonia
MRSA pneumonia is characterized by a massive influx of neutrophils into the lungs (26,27). To investigate a possible role for TLR9 signaling herein, we quantified neutrophils in BALF of tlr9−/− and Wt mice. Compared with Wt mice, tlr9−/− mice show a marked increase of neutrophil numbers in BALF harvested 24 and 48 h postinfection (p < 0.05, 24 h and p < 0.01, 48 h, respectively, Figure 4A). We wondered whether this enhanced neutrophil recruitment was related to elevated BALF levels of neutrophil attracting CXC chemokines. We therefore measured MIP-2 (Figure 4B) and KC (Figure 4C) in BALF obtained from both groups. MIP-2 tended to be higher in tir9−/− mice at all time points after infection significantly so at 72 h (p < 0.05 vs Wt mice). BALF KC levels were higher in tlr9−/− mice at 24 h (p < 0.01). Macrophage and lymphocyte counts in BALF increased over the course of the infection. After 48 h, tlr9r−/− mice showed a modest but significant increase in both macrophage and lymphocyte counts compared with Wt mice (Supplementary Table S1).
Enhanced neutrophil influx into the alveolar space of tlr9−/− mice during S. aureus pneumonia. Wt (gray) and tlr9−/− mice (white) were infected with 1 × 107 MRSA bacteria via intranasal inoculation and euthanized at the indicated time points thereafter. (A) Neutrophils, (B) MIP-2 and (C) KC in BALF are expressed as box-and-whisker diagrams depicting the median, the smallest observation, lower quartile, median, upper quartile and largest observation (n = 8 mice per group at each time point). *p < 0.05, **p < 0.01 versus Wt mice at the same time point.
TLR9 Deficiency Is Associated with Strongly Increased Lung Consolidation During MRSA Pneumonia
To evaluate the role of TLR9 in the severity of lung pathology, we analyzed pulmonary inflammation and injury in lung tissue slides obtained from tlr9−/− and Wt mice after infection with MRSA (Figure 5). After 24 h, all mice displayed signs of acute lung injury, including interstitial inflammation, endothelialitis, bronchitis and pleuritis; the extent of lung pathology upon histological examination remained relatively stable thereafter. Tlr9−/− and Wt mice displayed similar severities of lung pathology, as reflected by the semiquantitative scores composed as described in Materials and Methods, although at 48 h, tlr9−/− mice had modestly lower pathology scores versus Wt mice (p < 0.05, Figure 5F). Remarkably, tlr9−/− mice showed a strongly impaired resolution of lung inflammation, as reflected by large areas of lung consolidation at 24 and 48 h that were virtually absent in Wt mice (p < 0.05, Figure 5A, C, E). Detailed analyses of areas of lung consolidation indicated that more than 95% of the cells in these areas were neutrophils.
Tlr−/− mice show more consolidating pneumonia after induction of S. aureus pneumonia. Representative slides of lung HE staining of (A, B) Wt mice and (C, D) tlr9−/− mice. (A, C) depiction of a whole lung section; (B, D) ×200 magnification. (E) Consolidating pneumonia and (F) total pathology scores were determined at the indicated time points post infection in Wt mice (gray) and tlr9−/− mice (white) according to the scoring system described in the online supplement. Data are expressed as bar diagrams depicting mean ± standard error (n = 7–8 mice per group at each time point). *p < 0.05 versus Wt mice at the same time point.
Discussion
MRSA pneumonia has emerged as a major health care concern. MRSA infection of lower airways is feared for its association with massive cell death and necrosis, which is expected to result in the local release of DAMPs. We set out to determine whether one of these DAMPs, host DNA, can be detected in the bronchoalveolar space of mice infected with MRSA via the airways, and whether TLR9, one of the DNA sensors in mammalian cells, plays a role in the host response during MRSA pneumonia. We here demonstrate that MRSA elicits sustained release of host DNA in the lower respiratory tract, as reflected by high levels of nucleosomes and cell-free DNA in BALF. In addition, we show that another potential TLR9 ligand, bacterial DNA, is detected early after infection with MRSA, coinciding with high numbers of viable bacteria. TLR9 deficiency resulted in a delayed clearance of MRSA from the lungs, which was accompanied by increased neutrophil accumulation and markedly enhanced lung consolidation. These data suggest that the interaction between TLR9 and host- and pathogen-derived DNA has profound effects on the immune response during MRSA pneumonia.
Recent studies from our group reported the release of two other DAMPs in the bronchoalveolar space of mice infected with MRSA, that is, high-mobility group box 1 (HMGB1) (21) and myeloid-related protein (MRP8/14) (22). Although the appearance of MRP8/14 showed the same kinetics as detected for host DNA in the current investigation (22), HMGB1 demonstrated a transient increase in BALF peaking after 24 h (21). Nucleosomes are DNA segments wrapped around histone protein cores. Considering the nuclear origin of HMGB1, one might expect that the release of this protein follows similar kinetics as nucleosomes and DNA. Of note, however, HMGB1 can also be secreted actively (28); this, together with the possibility that nucleosomes, DNA and HMGB1 measured in BALF may at least in part originate from different cell types may explain the different kinetics of their release. Our study is limited by the fact that the cellular source of cell-free host DNA was not established. However, the marked increase in neutrophil counts and the similar kinetics of release of host DNA and MRP8/14 (22), which is mainly derived from neutrophils (29), suggest that neutrophils likely represent an important source of these DAMPs during MRSA pneumonia. Another likely source of released host DNA is the respiratory epithelium. Unlike host DNA, bacterial DNA especially was detected early after infection. We carefully removed viable bacteria from BALF by centrifugation prior to amplifying MRSA DNA in cell-free supernatants. We also demonstrated that these supernatants did not show growth of MRSA upon culture (data not shown), thereby excluding the possibility that we merely measured the bacterial load in these samples. As such, our data indicate that MRSA releases its DNA during growth, probably at least in part as a consequence of bacterial cell death.
TLR9 was the first PRR implicated in detection of DNA (30). In early reports, TLR9 was shown to recognize under-methylated DNA (CpG DNA), which is enriched in microbial genomes compared with mammalian cells. However, in later studies, TLR9 was also shown to be capable of sensing host DNA. For example, TLR9 deficiency reduced acetaminophen-induced liver injury and mortality by a mechanism that relied on attenuated DNA-induced inflammation (31). Similarly, in a heart failure mouse model, host DNA-TLR9 signaling caused inflammation and worsened cardiac dysfunction (32). Importantly, it is now clear that both self or pathogen DNA can activate multiple TLR9-independent pathways, including TANK-binding kinase-1, DAI (DNA-dependent activator of interferon regulatory transcription factors), STING (stimulator of interferon genes), AIM2 (absent in melanoma 2) and several others (14,15). Therefore, our investigation does not establish to which extent cell-free DNA contributes to the host response during MRSA pneumonia. In addition, we cannot be certain which DNA source (host or pathogen) drives the immune response in our model. Conceivably, the attenuated early release (6 h) of TNF-α and IL-6 in BALF in tlr9−/− mice may be partially explained by an absent interaction between TLR9 and either host or pathogen DNA, considering that both were present in the airways at this time point. IL-1β levels did not differ between mouse strains at 6 h after infection, which is consistent with the fact that the release of mature IL-1β requires activation of caspase-1 mediated by inflammasomes. In this respect, it should be noted that several DNA-sensing inflammasomes have been identified (33). At later time points after infection (especially 24 h), tlr9−/− mice had higher levels of IL-1β, IL-6, MIP-2 and KC in their BALF when compared with Wt mice, most likely, due to higher bacterial loads and activation of PRRs other than TLR9 by MRSA. Indeed, S. aureus can activate a variety of immune receptors, including TLR2 (34,35) and NOD2 (nucleotide-binding oligomerization domain-containing protein 2) (36,37). Increased BALF levels of IL-1β and chemokines likely contributed to the enhanced influx of neutrophils in tlr9−/− mice.
In our pneumonia model, MRSA disseminated to the liver while blood cultures remained sterile. This seeming discrepancy, also observed in our earlier studies (21,22), likely is explained by a more rapid clearance of this MRSA strain from the circulation relative to liver tissue. In accordance with this notion, very low bacterial burdens remain in the circulation upon intravenous administration of this MRSA strain with 100–1,000-fold higher bacterial loads in the liver (38).
Our data confirm and expand upon a previous study that examined the role of TLR9 in host defense during MRSA pneumonia (20). This former investigation reported reduced bacterial loads in lungs of tlr9−/− mice at a single time point (24 h) after infection, accompanied by lower TNF-α levels. The present report differs from this earlier one in that it studied the kinetics of the host reaction in more detail, providing more insight into both the early and the late inflammatory response, lung pathology and release of TLR9 ligands. The enhanced clearance of MRSA in tlr9−/− mice and the concomitantly reduced levels of TNF-α is in accordance with the negative impact of TNF signaling on bacterial loads during MRSA pneumonia (39). Moreover, previous studies also documented an association between reduced proinflammatory cytokine release into the airways and enhanced bacterial clearance during S. aureus pneumonia (40). Likewise, improved bacterial clearance was reported in tlr9−/− mice with polymicrobial abdominal sepsis (41). The advantageous role of TLR9 in bacterial clearance during respiratory tract infections is not unique for S. aureus: in models of pneumonia caused by Streptococcus pneumoniae (42), Klebsiella pneumoniae (43) or Legionella pneumophila (44), tlr9−/− mice also displayed increased bacterial burdens when compared with Wt mice, whereas TLR9 did not impact bacterial loads after airway infection with Haemophilus influenzae (45). Of considerable interest, tlr9−/− mice displayed a strongly enhanced lung consolidation during MRSA pneumonia, suggesting that TLR9 signaling contributes to the resolution of lung inflammation. In this respect, it is interesting to note that TLR signaling has been reported to contribute to the resolution of lung inflammation in noninfectious conditions (46,47).
Conclusion
MRSA can cause severe necrotizing pneumonia. The present study shows for the first time that experimentally induced respiratory tract infection with MRSA is associated with the release of both host- and pathogen-derived DNA in the airways. Genetic ablation of the DNA sensor TLR9 resulted in clear alterations in the host responses that involved a reduced early proinflammatory cytokine response accompanied by impaired bacterial clearance and a profound increase in lung consolidation. These results indicate that the interaction between cell-free DNA and TLR9 plays an important role in the innate immune response to MRSA in the lower airways.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Spellberg B, Bartlett JG, Gilbert DN. (2013) The future of antibiotics and resistance. N. Engl. J. Med. 368:299–302.
Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. (2006) Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 368:874–5.
DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. (2010) Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 375:1557–68.
Klevens RM, et al. (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 298:1763–71.
Kollef MH, et al. (2005) Epidemiology and outcomes of health-care-associated pneumonia: Results from a large US database of culture-positive pneumonia. Chest. 128:3854–62.
Defres S, Marwick C, Nathwani D. (2009) MRSA as a cause of lung infection including airway infection, community-acquired pneumonia and hospital-acquired pneumonia. Eur. Respir. J. 34:1470–6.
Kuehnert MJ, et al. (2006) Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J. Infect. Dis. 193:172–9.
Diep BA, Otto M. (2008) The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361–9.
Kono H, Rock KL. (2008) How dying cells alert the immune system to danger. Nat. Rev. Immunol. 8:279–89.
Chan JK, et al. (2012) Alarmins: Awaiting a clinical response. J. Clin. Invest. 122:2711–9.
Zeerleder S, et al. (2003) Elevated nucleosome levels in systemic inflammation and sepsis. Crit. Care Med. 31:1947–51.
Dwivedi DJ, et al. (2012) Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit. Care. 16:R151.
Forsblom E, et al. (2014) High cell-free DNA predicts fatal outcome among Staphylococcus aureus bacteraemia patients with intensive care unit treatment. PLoS One. 9:e87741.
Barber GN. (2011) Cytoplasmic DNA innate immune pathways. Immunol. Rev. 243:99–108.
Paludan S, Bowie A. (2013) Immune sensing of DNA. Immunity. 38:870–80.
Holm CK, Paludan SR, Fitzgerald KA. (2013) DNA recognition in immunity and disease. Curr. Opin. Immunol. 25:13–8.
Wagner H. (2001) Toll meets bacterial CpG-DNA. Immunity. 14:499–502.
Bauer S, et al. (2001) Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. U. S. A. 98:9237–42.
Hemmi H, et al. (2000) A Toll-like receptor recognizes bacterial DNA. Nature. 408:740–5.
Parker D, Prince A. (2012) Staphylococcus aureus induces type I IFN signaling in dendritic cells via TLR9. J. Immunol. 189:4040–6.
Achouiti A, et al. (2013) High-mobility group box 1 and the receptor for advanced glycation end products contribute to lung injury during Staphylococcus aureus pneumonia. Crit. Care. 17:R296.
Achouiti A, et al. (2015) Myeloid-related protein-14 deficiency promotes inflammation in staphylococcal pneumonia. Eur. Respir. J. 46:464–73.
Voyich JM, et al. (2006) Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–70.
van Nieuwenhuijze AE, van Lopik T, Smeenk RJ, Aarden LA. (2003) Time between onset of apoptosis and release of nucleosomes from apoptotic cells: Putative implications for systemic lupus erythematosus. Ann. Rheum. Dis. 62:10–4.
Zeerleder S, et al. (2012) Circulating nucleosomes and severity of illness in children suffering from meningococcal sepsis treated with protein C. Crit. Care Med. 40:3224–9.
Craig A, Mai J, Cai S, Jeyaseelan S. (2009) Neutrophil recruitment to the lungs during bacterial pneumonia. Infect. Immun. 77:568–75.
Bartlett AH, Foster TJ, Hayashida A, Park PW. (2008) Alpha-toxin facilitates the generation of CXC chemokine gradients and stimulates neutrophil homing in Staphylococcus aureus pneumonia. J. Infect. Dis. 198:1529–35.
Lu B, et al. (2014) Molecular mechanism and therapeutic modulation of high mobility group box 1 release and action: An updated review. Expert. Rev. Clin. Immunol. 10:713–27.
Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. (2009) The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 86:557–66.
Dempsey A, Bowie AG. (2015) Innate immune recognition of DNA: A recent history. Virology. 479-80:146–52.
Imaeda AB, et al. (2009) Acetaminophen-induced hepatotoxicity in mice is dependent on Tlr9 and the Nalp3 inflammasome. J. Clin. Invest. 119:305–14.
Oka T, et al. (2012) Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 485:251–5.
Choubey D. (2012) DNA-responsive inflammasomes and their regulators in autoimmunity. Clin. Immunol. 142:223–31.
Schmaler M, et al. (2009) Lipoproteins in Staphylococcus aureus mediate inflammation by TLR2 and iron-dependent growth in vivo. J. Immunol. 182:7110–8.
Gillrie MR, et al. (2010) Divergent roles of Tolllike receptor 2 in response to lipoteichoic acid and Staphylococcus aureus in vivo. Eur. J. Immunol. 40:1639–50.
Girardin SE, et al. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869–72.
Deshmukh HS, et al. (2009) Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect. Immun. 77:1376–82.
Achouiti A, Van’t Veer C, de Vos AF, van der Poll T. (2015) The receptor for advanced glycation end products promotes bacterial growth at distant body sites in Staphylococcus aureus skin infection. Microbes Infect. 17:622–7.
Gomez MI, et al. (2004) Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842–8.
Parker D, Prince A. (2011) Immunopathogenesis of Staphylococcus aureus pulmonary infection. Semin. Immunopathol. 34:281–97.
Plitas G, Burt BM, Nguyen HM, Bamboad ZM, DeMatteo RP. (2008) Toll-like receptor 9 inhibition reduces mortality in polymicrobial sepsis. J. Exp. Med. 205:1277–83.
Albiger B, et al. (2007) Toll-like receptor 9 acts at an early stage in host defence against pneumococcal infection. Cell Microbiol. 9:633–44.
Bhan U, et al. (2007) TLR9 is required for protective innate immunity in Gram-negative bacterial pneumonia: Role of dendritic cells. J. Immunol. 179:3937–46.
Bhan U, et al. (2008) Toll-like receptor 9 regulates the lung macrophage phenotype and host immunity in murine pneumonia caused by Legionella pneumophila. Infect. Immun. 76:2895–904.
Wieland CW, Florquin S, van der Poll T. (2010) Toll-like receptor 9 is not important for host defense against Haemophilus influenzae. Immunobiology. 215:910–4.
Jiang D, Liang J, Li Y, Noble PW. (2006) The role of Toll-like receptors in non-infectious lung injury. Cell Res. 16:693–701.
Yang H-Z, et al. (2012) TLR4 activity is required in the resolution of pulmonary inflammation and fibrosis after acute and chronic lung injury. Am. J. Pathol. 180:275–92.
Acknowledgments
We thank Regina de Beer, Joost Daalhuisen and Marieke S. ten Brink for expert technical assistance. We would like to thank Dr. Ellen van der Schoot for providing equipment for setting up the bacterial DNA assay and Dr. Shizuo Akira (Research Institute for Microbial Disease, Osaka, Japan) for generously providing us with tlr9−/− mice. We thank Dr. Timothy J. Foster (Microbiology Department, Trinity College, Dublin, Ireland) for the S. aureus strain used in these experiments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
10020_2016_2201292_MOESM1_ESM.pdf
Toll-Like Receptor 9 Enhances Bacterial Clearance and Limits Lung Consolidation in Murine Pneumonia Caused by Methicillin-Resistant Staphylococcus aureus
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this license, visit (http://creativecommons.org/licenses/by-nc-nd/4.0/)
About this article
Cite this article
van der Meer, A.J., Achouiti, A., van der Ende, A. et al. Toll-Like Receptor 9 Enhances Bacterial Clearance and Limits Lung Consolidation in Murine Pneumonia Caused by Methicillin-Resistant Staphylococcus aureus. Mol Med 22, 292–299 (2016). https://doi.org/10.2119/molmed.2015.00242
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2015.00242