Abstract
Background
Circular RNAs (circRNAs) have recently emerged as novel non-coding regulatory RNAs, reportedly involved in many biological processes and human diseases. However, the role of circRNAs in the pathogenesis of psoriasis is unclear. Additionally, mesenchymal stem cells (MSCs) are involved in pathological processes associated with immune diseases, including psoriasis.
Objectives
In this study, we determined the circRNA expression profile of MSCs from psoriatic skin lesions and investigated possible mechanisms associated with psoriasis.
Materials & Methods
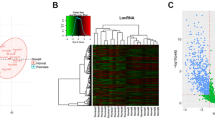
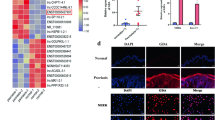
RNA sequencing was used to detect circRNA expression in MSCs from psoriatic skin lesions and normal skin, and detailed bioinformatic analysis was performed. Our results allowed us to predict a circRNA-microRNA (miRNA) interaction network and subsequently validate seven differentially expressed circRNAs and three miRNAs by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). Furthermore, we analysed circRNA expression in plasma to determine the extent of abnormal circRNA circulation in patients with psoriasis.
Results
In total, 6,323 circRNAs were detected, of which 3,227 constituted previously unreported circRNAs. We identified 129 circRNAs that exhibited significantly different expression patterns between the psoriasis and control groups, and established a circRNA-miRNA interaction network through bioinformatic analysis that indicated circRNA interaction with miRNAs associated with psoriasis. Additionally, qRT-PCR results confirmed the differential expression of seven circRNAs as well as the stable expression of three circRNAs in plasma from psoriasis patients, consistent with trends observed in cells.
Conclusions
This is the first study of circRNA expression in psoriasis, indicating possible circRNA involvement in the pathogenesis of psoriasis.
Similar content being viewed by others
References
Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15: 409.
Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015; 5: 472–80.
Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA 2010; 16: 2043–50.
Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol 2010; 20: R858–61.
Elder JT, Bruce AT, Gudjonsson JE, et al. Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol 2010; 130: 1213–26.
Nograles KE, Davidovici B, Krueger JG. New insights in the immunologic basis of psoriasis. Semin Cutan Med Surg 2010; 29: 3–9.
Wolf R, Mascia F, Dharamsi A, et al. Gene from a psoriasis susceptibility locus primes the skin for inflammation. Sci Transl Med 2010; 2: 61ra90.
Huang RY, Li L, Wang MJ, Chen XM, Huang QC, Lu CJ. An exploration of the role of microRNAs in psoriasis: a systematic review of the literature. Medicine (Baltimore) 2015; 94: e2030.
Gulati N, Løvendorf MB, Zibert JR, et al. Unique microRNAs appear at different times during the course of a delayed-type hypersensitivity reaction in human skin. Exp Dermatol 2015; 24: 953–7.
Zhang W, Yi X, Guo S, et al. A single-nucleotide polymorphism of miR-146a and psoriasis: an association and functional study. J Cell Mol Med 2014; 18: 2225–34.
Jiang S, Hinchliffe TE, Wu T. Biomarkers of an autoimmune skin disease-psoriasis. Genomics Proteomics Bioinformatics 2015; 13: 224–33.
Hou R, Yin G, An P, et al. DNA methylation of dermal MSCs in psoriasis: identification of epigenetically dysregulated genes. J Dermatol Sci 2013; 72: 103–9.
Liu R, Yang Y, Yan X, Zhang K. Abnormalities in cytokine secretion from mesenchymal stem cells in psoriatic skin lesions. Eur J Dermatol 2013; 23: 600–7.
Liu R, Wang Y, Zhao X, Yang Y, Zhang K. Lymphocyte inhibition is compromised in mesenchymal stem cells from psoriatic skin. Eur J Dermatol 2014; 24: 560–7.
Campanati A, Orciani M, Consales V, et al. Characterization and profiling of immunomodulatory genes in resident mesenchymal stem cells reflect the Th1-Th17/Th2 imbalance of psoriasis. Arch Dermatol Res 2014; 306: 915–20.
Orciani M, Campanati A, Salvolini E, et al. The mesenchymal stem cell profile in psoriasis. Br J Dermatol 2011; 165: 585–92.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics 2013; 1303: 3997.
Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015; 16: 4.
Meng X, Chen Q, Zhang P, Chen M. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics 2017; 33: 3314–6.
Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 2016; 7: 12060.
Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20: 1666–70.
Xing Z, Chu C, Chen L, Kong X. The use of gene ontology terms and KEGG pathways for analysis and prediction of oncogenes. B iochim Biophys Acta 2016; 1860: 2725–34.
Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 2016; 428: 726–31.
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92–105.
Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010; 11: R90.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011; 27: 431–2.
Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006; 8: 315–7.
Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25: 981–4.
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6: e1001233.
Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015; 5: 12453.
Yin X, Low HQ, Wang L, et al. Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat Commun 2015; 6: 6916.
Swindell WR, Stuart PE, Sarkar MK, et al. Cellular dissection of psoriasis for transcriptome analyses and the post-GWAS era. BMC Med Genomics 2014; 7: 27.
Qi M, Elion EA. MAP kinase pathways. J Cell Sci 2005; 118: 3569–72.
Mavropoulos A, Rigopoulou EI, Liaskos C, Bogdanos DP, Sakkas LI. The role of p38 MAPK in the aetiopathogenesis of psoriasis and psoriatic arthritis. Clin Dev Immunol 2013; 2013: 569751.
He Q, Chen HX, Li W, et al. IL-36 cytokine expression and its relationship with p38 MAPK and NF-κB pathways in psoriasis vulgaris skin lesions. J Huazhong Univ Sci Technolog Med Sci 2013; 33: 594–9.
Johansen C, Funding AT, Otkjaer K, et al. Protein expression of TNF-alpha in psoriatic skin is regulated at a posttranscriptional level by MAPK-activated protein kinase 2. J Immunol 2006; 176: 1431–8.
Dimon-Gadal S, Raynaud F, Evain-Brion D, et al. MAP kinase abnormalities in hyperproliferative cultured fibroblasts from psoriatic skin. J Invest Dermatol 1998; 110: 872–9.
Haase I, Hobbs RM, Romero MR, Broad S, Watt FM. A role for mitogen-activated protein kinase activation by integrins in the pathogenesis of psoriasis. J Clin Invest 2001; 108: 527–36.
Joyce CE, Zhou X, Xia J, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 2011; 20: 4025–40.
Guo S, Zhang W, Wei C, et al. Serum and skin levels of miR-3693p in patients with psoriasis and their correlation with disease severity. Eur J Dermatol 2013; 23: 608–13.
Hawkes JE, Nguyen GH, Fujita M, et al. microRNAs in psoriasis. J Invest Dermatol 2016; 136: 365–71.
Griffiths CE, Barker JN. Pathogenesis and clinical features of psoriasis. Lancet 2007; 370: 263–71.
Schon MP, Boehncke WH. Psoriasis. N Engl J Med 2005; 352: 1899–912.
Hong J, Koo B, Koo J. The psychosocial and occupational impact of chronic skin disease. Dermatol Ther 2008; 21: 54–9.
Zachariae R, Zachariae H, Blomqvist K, et al. Quality of life in 6497 Nordic patients with psoriasis. Br J Dermatol 2002; 146: 1006–16.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Liu, R., Wang, Q., Chang, W. et al. Characterisation of the circular RNA landscape in mesenchymal stem cells from psoriatic skin lesions. Eur J Dermatol 29, 29–38 (2019). https://doi.org/10.1684/ejd.2018.3483
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1684/ejd.2018.3483