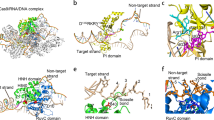
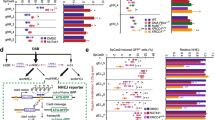
Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) is widely used for targeted genomic and epigenomic modifications and imaging in cells and organisms, and holds tremendous promise in clinical applications. The efficiency and accuracy of the technology are partly determined by the target binding affinity and residence time of Cas9-single-guide RNA (sgRNA) at a given site. However, little attention has been paid to the effect of target binding affinity and residence duration on the repair of Cas9-induced DNA double-strand breaks (DSBs). We propose that the choice of DSB repair pathway may be altered by variation in the binding affinity and residence duration of Cas9-sgRNA at the cleaved target, contributing to significantly heterogeneous mutations in CRISPR/Cas9 genome editing. Here, we discuss the effect of Cas9-sgRNA target binding and residence on the choice of DSB repair pathway in CRISPR/Cas9 genome editing, and the opportunity this presents to optimize Cas9-based technology.
摘要
CRISPR/Cas9技术广泛应用于靶向基因编辑、表观遗传学修饰和细胞成像等多个领域,临床应用潜能巨大。然而,Cas9-sgRNA复合物靶点结合强度与滞留时间长短是否会对Cas9诱导的DNA双链断裂(DSB)修复产生影响并不清楚,这个问题也常被忽视。我们先前的研究发现,CRISPR/Cas9技术的有效性和精准性部分取决于Cas9-sgRNA在靶点的结合以及滞留,其靶点结合亲和力和滞留时间会随着靶点不同而不同,从而影响DSB修复途径的选择,这也是CRISPR/Cas9基因编辑异质性产生的一个重要原因。在本文中,我们将讨论CRISPR/Cas9基因编辑中Cas9-sgRNA的靶点结合与滞留如何影响细胞内DSB修复途径的选择,在此基础上提出优化CRISPR/Cas9技术的可能方式。
Similar content being viewed by others
Change history
08 April 2021
The typesetting format of the online version of the first issue (2021 22(01)) of Journal of Zhejiang University-SCIENCE B is different from that of the printed version (but all the text, figure and table contents in the article are correct). This is due to the new typesetting company adopted this year.
References
Abadi S, Yan WX, Amar D, et al., 2017. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol, 13(10):e1005807. https://doi.org/10.1371/journal.pcbi.1005807
Allen F, Crepaldi L, Alsinet C, et al., 2019. Predicting the mutations generated by repair of Cas9-induced doublestrand breaks. Nat Biotechnol, 37(1):64–72. https://doi.org/10.1038/nbt.4317
Anders C, Niewoehner O, Duerst A, et al., 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature, 513(7519):569–573. https://doi.org/10.1038/nature13579
Bakkenist CJ, Kastan MB, 2015. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair, 36:8–12. https://doi.org/10.1016/j.dnarep.2015.09.002
Bhargava R, Onyango DO, Stark JM, 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet, 32(9):566–575. https://doi.org/10.1016/j.tig.2016.06.007
Bisaria N, Jarmoskaite I, Herschlag D, 2017. Lessons from enzyme kinetics reveal specificity principles for RNA-guided nucleases in RNA interference and CRISPR-based genome editing. Cell Syst, 4(1):21–29. https://doi.org/10.1016/j.cels.2016.12.010
Blackford AN, Jackson SP, 2017. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell, 66(6):801–817. https://doi.org/10.1016/j.molcel.2017.05.015
Boboila C, Alt FW, Schwer B, 2012. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol, 116:1–49. https://doi.org/10.1016/B978-0-12-394300-2.00001-6
Bolukbasi MF, Gupta A, Oikemus S, et al., 2015. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods, 12(12): 1150–1156. https://doi.org/10.1038/nmeth.3624
Boyle EA, Andreasson JOL, Chircus LM, et al., 2017. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci USA, 114(21):5461–5466. https://doi.org/10.1073/pnas.1700557114
Bustamante C, Bryant Z, Smith SB, 2003. Ten years of tension: single-molecule DNA mechanics. Nature, 421(6921):423–427. https://doi.org/10.1038/nature01405
Casini A, Olivieri M, Petris G, et al., 2018. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol, 36(3):265–271. https://doi.org/10.1038/nbt.4066
Ceccaldi R, Sarangi P, D’Andrea AD, 2016. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol, 17(6):337–349. https://doi.org/10.1038/nrm.2016.48
Chakrabarti AM, Henser-Brownhill T, Monserrat J, et al., 2019. Target-specific precision of CRISPR-mediated genome editing. Mol Cell, 73(4):699–713.e6. https://doi.org/10.1016/j.molcel.2018.11.031
Chang HHY, Watanabe G, Gerodimos CA, et al., 2016. Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. J Biol Chem, 291(47):24377–24389. https://doi.org/10.1074/jbc.M116.752329
Chen BH, Gilbert LA, Cimini BA, et al., 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 155(7):1479–1491. https://doi.org/10.1016/j.cell.2013.12.001
Chen JS, Dagdas YS, Kleinstiver BP, et al., 2017. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature, 550(7676):407–410. https://doi.org/10.1038/nature24268
Chu VT, Weber T, Wefers B, et al., 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol, 33(5):543–548. https://doi.org/10.1038/nbt.3198
Chuai GH, Ma HH, Yan JF, et al., 2018. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol, 19:80. https://doi.org/10.1186/s13059-018-1459-4
Ciccia A, Elledge SJ, 2010. The DNA damage response: making it safe to play with knives. Mol Cell, 40(2):179–204. https://doi.org/10.1016/j.molcel.2010.09.019
Clarke R, Heler R, MacDougall MS, et al., 2018. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol Cell, 71(1):42–55.e8. https://doi.org/10.1016/j.molcel.2018.06.005
Cong L, Ran FA, Cox D, et al., 2013. Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121): 819–823. https://doi.org/10.1126/science.1231143
Doench JG, Fusi N, Sullender M, et al., 2016. Optimized sgRNA design to maximize activity and minimize offtarget effects of CRISPR-Cas9. Nat Biotechnol, 34(2): 184–191. https://doi.org/10.1038/nbt.3437
Feng YL, Xiang JF, Kong N, et al., 2016. Buried territories: heterochromatic response to DNA double-strand breaks. Acta Biochim Biophys Sin (Shanghai), 48(7):594–602. https://doi.org/10.1093/abbs/gmw033
Feng YL, Xiang JF, Liu SC, et al., 2017. H2AX facilitates classical non-homologous end joining at the expense of limited nucleotide loss at repair junctions. Nucleic Acids Res, 45(18):10614–10633. https://doi.org/10.1093/nar/gkx715
Fu YF, Sander JD, Reyon D, et al., 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol, 32(3):279–284. https://doi.org/10.1038/nbt.2808
Gallagher DN, Haber JE, 2018. Repair of a site-specific DNA cleavage: old-school lessons for Cas9-mediated gene editing. ACS Chem Biol, 13(2):397–405. https://doi.org/10.1021/acschembio.7b00760
Garneau JE, Dupuis MÈ, Villion M, et al., 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature, 468(7320):67–71. https://doi.org/10.1038/nature09523
Gaudelli NM, Komor AC, Rees HA, et al., 2017. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551(7681):464–471. https://doi.org/10.1038/nature24644
Gilbert LA, Larson MH, Morsut L, et al., 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154(2):442–451. https://doi.org/10.1016/j.cell.2013.06.044
Guo T, Feng YL, Xiao JJ, et al., 2018. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol, 19:170. https://doi.org/10.1186/s13059-018-1518-x
Hegazy YA, Fernando CM, Tran EJ, 2020. The balancing act of R-loop biology: the good, the bad, and the ugly. J Biol Chem, 295(4):905–913. https://doi.org/10.1074/jbc.REV119.011353
Hilton IB, D’Ippolito AM, Vockley CM, et al., 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol, 33(5):510–517. https://doi.org/10.1038/nbt.3199
Hinz JM, Laughery MF, Wyrick JJ, 2015. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry, 54(48): 7063–7066. https://doi.org/10.1021/acs.biochem.5b01108
Horvath P, Barrangou R, 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science, 327(5962):167–170. https://doi.org/10.1126/science.1179555
Hsu PD, Lander ES, Zhang F, 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157(6):1262–1278. https://doi.org/10.1016/j.cell.2014.05.010
Hu JH, Miller SM, Geurts MH, et al., 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature, 556(7699):57–63. https://doi.org/10.1038/nature26155
Isaac RS, Jiang FG, Doudna JA, et al., 2016. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife, 5:e13450. https://doi.org/10.7554/eLife.13450
Ivanov IE, Wright AV, Cofsky JC, et al., 2020. Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc Natl Acad Sci USA, 117(11):5853–5860. https://doi.org/10.1073/pnas.1913445117
Jasin M, Rothstein R, 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol, 5(11):a012740. https://doi.org/10.1101/cshperspect.a012740
Jasin M, Haber JE, 2016. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair, 44:6–16. https://doi.org/10.1016/j.dnarep.2016.05.001
Jeon Y, Choi YH, Jang YS, et al., 2018. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat Commun, 9:2777. https://doi.org/10.1038/s41467-018-05245-x
Jiang FG, Doudna JA, 2017. CRISPR-Cas9 structures and mechanisms. Ann Rev Biophys, 46:505–529. https://doi.org/10.1146/annurev-biophys-062215-010822
Jinek M, Chylinski K, Fonfara I, et al., 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096):816–821. https://doi.org/10.1126/science.1225829
Jones DL, Leroy P, Unoson C, et al., 2017. Kinetics of dCas9 target search in Escherichia coli. Science, 357(6358): 1420–1424. https://doi.org/10.1126/science.aah7084
Kearns NA, Pham H, Tabak B, et al., 2015. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods, 12(5):401–403. https://doi.org/10.1038/nmeth.3325
Kim D, Luk K, Wolfe SA, et al., 2019. Evaluating and enhancing target specificity of gene-editing nucleases and deaminases. Annu Rev Biochem, 88:191–220. https://doi.org/10.1146/annurev-biochem-013118-111730
Kim S, Kim D, Cho SW, et al., 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res, 24(6): 1012–1019. https://doi.org/10.1101/gr.171322.113
Kleinstiver BP, Prew MS, Tsai SQ, et al., 2015a. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol, 33(12):1293–1298. https://doi.org/10.1038/nbt.3404
Kleinstiver BP, Prew MS, Tsai SQ, et al., 2015b. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature, 523(7561):481–485. https://doi.org/10.1038/nature14592
Kleinstiver BP, Pattanayak V, Prew MS, et al., 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529(7587):490–495. https://doi.org/10.1038/nature16526
Knight SC, Xie LQ, Deng WL, et al., 2015. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science, 350(6262):823–826. https://doi.org/10.1126/science.aac6572
Komor AC, Kim YB, Packer MS, et al., 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533(7603):420–424. https://doi.org/10.1038/nature17946
Lemos BR, Kaplan AC, Bae JE, et al., 2018. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci USA, 115(9):E2040–E2047. https://doi.org/10.1073/pnas.1716855115
Lieber MR, 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem, 79:181–211. https://doi.org/10.1146/annurev.biochem.052308.093131
Lin S, Staahl BT, Alla RK, et al., 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife, 3:e04766. https://doi.org/10.7554/eLife.04766
Ma HH, Naseri A, Reyes-Gutierrez P, et al., 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA, 112(10):3002–3007. https://doi.org/10.1073/pnas.1420024112
Ma HH, Tu LC, Naseri A, et al., 2016. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J Cell Biol, 214(5):529–537. https://doi.org/10.1083/jcb.201604115
Mali P, Yang LH, Esvelt KM, et al., 2013. RNA-guided human genome engineering via Cas9. Science, 339(6121):823–826. https://doi.org/10.1126/science.1232033
Maruyama T, Dougan SK, Truttmann MC, et al., 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol, 33(5):538–542. https://doi.org/10.1038/nbt.3190
Newton MD, Taylor BJ, Driessen RPC, et al., 2019. DNA stretching induces Cas9 off-target activity. Nat Struct Mol Biol, 26(3):185–192. https://doi.org/10.1038/s41594-019-0188-z
Nishimasu H, Ran FA, Hsu PD, et al., 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156(5):935–949. https://doi.org/10.1016/j.cell.2014.02.001
Ouyang J, Lan L, Zou L, 2017. Regulation of DNA break repair by transcription and RNA. Sci China Life Sci, 60(10): 1081–1086. https://doi.org/10.1007/s11427-017-9164-1
Patel SS, Pandey M, Nandakumar D, 2011. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr Opin Chem Biol, 15(5):595–605. https://doi.org/10.1016/j.cbpa.2011.08.003
Perez-Pinera P, Kocak DD, Vockley CM, et al., 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods, 10(10):973–976. https://doi.org/10.1038/nmeth.2600
Puget N, Miller KM, Legube G, 2019. Non-canonical DNA/RNA structures during transcription-coupled doublestrand break repair: roadblocks or Bona fide repair intermediates? DNA Repair, 81:102661. https://doi.org/10.1016/j.dnarep.2019.102661
Qi LS, Larson MH, Gilbert LA, et al., 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152(5): 1173–1183. https://doi.org/10.1016/j.cell.2013.02.022
Ran FA, Hsu PD, Lin CY, et al., 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 154(6):1380–1389. https://doi.org/10.1016/j.cell.2013.08.021
Rees HA, Liu DR, 2018. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet, 19(12):770–788. https://doi.org/10.1038/s41576-018-0059-1
Richardson CD, Ray GJ, DeWitt MA, et al., 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol, 34(3):339–344. https://doi.org/10.1038/nbt.3481
Richardson CD, Kazane KR, Feng SJ, et al., 2018. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet, 50(8):1132–1139. https://doi.org/10.1038/s41588-018-0174-0
Scully R, Panday A, Elango R, et al., 2019. DNA doublestrand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 20(11):698–714. https://doi.org/10.1038/s41580-019-0152-0
Seol JH, Shim EY, Lee SE, 2018. Microhomology-mediated endjoining: good, bad and ugly. Mutat Res, 809:81–87. https://doi.org/10.1016/j.mrfmmm.2017.07.002
Shou J, Li JH, Liu YB, et al., 2018. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 71(4): 498–509.e4. https://doi.org/10.1016/j.molcel.2018.06.021
Singh D, Mallon J, Poddar A, et al., 2018. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc Natl Acad Sci USA, 115(21):5444–5449. https://doi.org/10.1073/pnas.1718686115
Slaymaker IM, Gao LY, Zetsche B, et al., 2016. Rationally engineered Cas9 nucleases with improved specificity. Science, 351(6268):84–88. https://doi.org/10.1126/science.aad5227
Sternberg SH, Redding S, Jinek M, et al., 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature, 507(7490):62–67. https://doi.org/10.1038/nature13011
Sternberg SH, LaFrance B, Kaplan M, et al., 2015. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature, 527(7576):110–113. https://doi.org/10.1038/nature15544
Strohkendl I, Saifuddin FA, Rybarski JR, et al., 2018. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol Cell, 71(5):816–824.e3. https://doi.org/10.1016/j.molcel.2018.06.043
Symington LS, Gautier J, 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet, 45: 247–271. https://doi.org/10.1146/annurev-genet-110410-132435
Szczelkun MD, Tikhomirova MS, Sinkunas T, et al., 2014. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA, 111(27):9798–9803. https://doi.org/10.1073/pnas.1402597111
Tanaka H, Yao MC, 2009. Palindromic gene amplification—an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer, 9(3):216–224. https://doi.org/10.1038/nrc2591
Verkuijl SAN, Rots MG, 2019. The influence of eukaryotic chromatin state on CRISPR-Cas9 editing efficiencies. Curr Opin Biotechnol, 55:68–73. https://doi.org/10.1016/j.copbio.2018.07.005
Wang HF, la Russa M, Qi LS, 2016. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem, 85:227–264. https://doi.org/10.1146/annurev-biochem-060815-014607
Yeh CD, Richardson CD, Corn JE, 2019. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol, 21(12):1468–1478. https://doi.org/10.1038/s41556-019-0425-z
Zetsche B, Gootenberg JS, Abudayyeh OO, et al., 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163(3):759–771. https://doi.org/10.1016/j.cell.2015.09.038
Zhang Q, Wen FC, Zhang SQ, et al., 2019. The post-PAM interaction of RNA-guided spCas9 with DNA dictates its target binding and dissociation. Sci Adv, 5(11):eaaw9807. https://doi.org/10.1126/sciadv.aaw9807
Zhang SQ, Zhang Q, Hou XM, et al., 2020. Dynamics of Staphylococcus aureus Cas9 in DNA target association and dissociation. EMBO Rep, 21:e50184. https://doi.org/10.15252/embr.202050184
Zhang XH, Chen L, Zhu BY, et al., 2020. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat Cell Biol, 22(6):740–750. https://doi.org/10.1038/s41556-020-0518-8
Zhang YX, Pan WY, Chen J, 2019. p53 and its isoforms in DNA double-stranded break repair. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 20(6):457–466. https://doi.org/10.1631/jzus.B1900167
Zhu X, Clarke R, Puppala AK, et al., 2019. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol, 26(8):679–685. https://doi.org/10.1038/s41594-019-0258-2
Acknowledgments
The research is supported by the National Natural Science Foundation of China (Nos. 31671385 and 31870806), the Zhejiang Provincial Natural Science Foundation of China (Nos. LY18C050001 and LQ20C050004), and the Fundamental Research Funds for the Central Universities in China (No. 2019QNA7031). We thank members of the XIE’s lab for their contributions and helpful discussions. We sincerely apologize to authors who are not quoted in references due to space limitations.
Author information
Authors and Affiliations
Contributions
Anyong XIE, Yili FENG, and Sicheng LIU wrote the manuscript. Yili FENG drew the figures. Anyong XIE, Yili FENG, Sicheng LIU, and Ruodan CHEN edited the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Additional information
Compliance with ethics guidelines
Yili FENG, Sicheng LIU, Ruodan CHEN, and Anyong XIE declare that they have no conflicts of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
Rights and permissions
About this article
Cite this article
Feng, Y., Liu, S., Chen, R. et al. Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing. J. Zhejiang Univ. Sci. B 22, 73–86 (2021). https://doi.org/10.1631/jzus.B2000282
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B2000282
Key words
- CRISPR/Cas9 genome editing
- Double-strand break (DSB) repair pathway choice
- Target binding affinity
- Target residence