Abstract
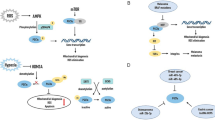
Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) is a transcriptional co-activator involved in mitochondrial biogenesis, respiratory capacity, and oxidative phosphorylation (OXPHOS). PGC-1α plays an important role in cellular metabolism and is associated with tumorigenesis, suggesting an involvement in cell cycle progression. However, the underlying mechanisms mediating its involvement in these processes remain unclear. To elucidate the signaling pathways involved in PGC-1α function, we established a cell line, CH1 PGC-1α, which stably overexpresses PGC-1α. Using this cell line, we found that over-expression of PGC-1α stimulated extra adenosine triphosphate (ATP) and reduced reactive oxygen species (ROS) production. These effects were accompanied by up-regulation of the cell cycle checkpoint regulators CyclinD1 and CyclinB1. We hypothesized that ATP and ROS function as cellular signals to regulate cyclins and control cell cycle progression. Indeed, we found that reduction of ATP levels down-regulated CyclinD1 but not CyclinB1, whereas elevation of ROS levels down-regulated CyclinB1 but not CyclinD1. Furthermore, both low ATP levels and elevated ROS levels inhibited cell growth, but PGC-1α was maintained at a constant level. Together, these results demonstrate that PGC-1α regulates cell cycle progression through modulation of CyclinD1 and CyclinB1 by ATP and ROS. These findings suggest that PGC-1α potentially coordinates energy metabolism together with the cell cycle.
中文概要
目 的
探讨在CH1 细胞中过氧化物酶体增殖物受体γ 共激活因子1α(PGC-1α)调控细胞周期时三磷 酸腺苷(ATP)和活性氧(ROS)的作用机制。
创新点
构建了稳定表达PGC-1α 的CH1 细胞株, 并系统 地研究了PGC-1α 调控细胞周期是通过ATP 和 ROS 调节CyclinD1 和CyclinB1 的行使功能。
方 法
以慢病毒质粒pBABE 为载体构建了PGC-1α 稳 定表达的CH1 PGC-1α 细胞株(PGC-1α), 同时 转染空质粒pBABE 作为对照(PB), 结合RNA 干扰CH1 PGC-1α 中PGC-1α 的过表达(Si), 测定了ATP 和ROS 水平。用流式细胞术检测了 细胞周期和免疫印迹检测了CyclinB1/D1 的表达, 并进一步分别用寡霉素抑制PGC-1α 细胞中的 ATP 生成, 用H2O2 处理细胞以增加外源ROS 水 平。然后检测ATP 和ROS 改变后, 对CyclinB1/D1 表达及细胞周期的影响, 以明确ATP 和ROS 是 否参与PGC-1α 对细胞周期的调控作用。
结 论
本实验成功构建了稳定表达PGC-1α 的细胞株 (图1 和图2a), 与PB 对照和RNA 干扰PGC-1α 比较, 过表达PGC-1α 具有升高ATP、降低ROS 和促进细胞周期的作用(图3 和图4)。进一步 用寡霉素抑制ATP 合成后发现CyclinD1 明显下 调(图5), 而加入H2O2 增加外源ROS 后发现 CyclinB1 显著上调(图6)。通过本实验我们提 出PGC-1α调控细胞周期是通过升高ATP水平抑 制CyclinD1 表达和降低ROS 水平促进CyclinB1 表达来实现。
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Impairment of mitochondrial oxidative phosphorylation (OXPHOS) leads to depletion of adenosine triphosphate (ATP) and overproduction of reactive oxygen species (ROS), which further induce cell growth defects in mitochondrial disease (Wallace, 2005; Chaturvedi and Beal, 2013). The transcriptional co-activator peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) is a key regulator of signaling pathways that promote mitochondrial biogenesis and bioenergetics in cardiomyocytes, and PGC-1α regulates peroxisome proliferator-activated receptor α (PPARα) to enhance the transcription of mitochondrial fatty acid β-oxidation enzymes (Wu et al., 1999; Lehman et al., 2000; Vega et al., 2000). It interacts with a broad range of transcription factors, including nuclear hormone receptors, nuclear respiratory factors, and specific transcription factors, to regulate glucose metabolism and response to environmental stimuli such as cold exposure and prolonged starvation (Cannon et al., 1998; Meirhaeghe et al., 2003; Lin et al., 2005). Consistent with these roles, it is expressed at high levels in tissues with a high energy demand, such as brain, kidney, cardiac, and skeletal muscle (Lin et al., 2005). Therefore, PGC-1α dysregulation has critical roles in vascular disease, diabetes, and neurodegenerative disease (Lin et al., 2005). The mechanisms governing cellular energy homeostasis have been an area of active investigation, and numerous studies have implicated PGC-1α as a key player in the control of energy homeostasis (Rohas et al., 2007). However, the mechanisms by which PGC-1α exerts this control have remained unclear. Mitochondria are essential for producing the energy source ATP as well as ROS by-products. ATP generation is finely tuned to match energy demand as an important signal for macromolecule production and DNA synthesis (McBride et al., 2006). Tumor cells, for example, require high levels of ATP for survival, proliferation, and metastasis (Weinberg and Chandel, 2009). Dysfunctional mitochondria are associated with reduced levels of ATP and excessive production of ROS (Pieczenik and Neustadt, 2007). The understanding of ATP and ROS provides significant insight to possible treatments for mitochondrial diseases.
Mitochondrial biogenesis is essential to cell division. It cannot be produced spontaneously and must be inherited at cell division (Shiota et al., 2015). In eukaryotes, the cell cycle is a sequential series of events with checkpoints at the transition from G1 to S phase (G1/S) and at the transition from G2 to M phase (G2/M) (Löbrich and Jeggo, 2007; Bertoli et al., 2013). CyclinD and CyclinB are members of the cyclin protein family, and interact with cyclin-dependent kinases (CDKs) to regulate the two checkpoints (Malumbres and Barbacid, 2009). Although mitochondrial dysfunction and oxidative stress are heavily involved in various cardiovascular and neurodegenerative diseases, their roles in the cell cycle remain unknown. We hypothesized that mitochondrial OXPHOS capacity is the main driving force of cell cycle progression.
Our work focused on the effects of reverting the PGC-1α function (into excessive oxidative stress and bioenergetic deficits) on cell cycle, and our aim was to reveal the signaling pathways mediating mitochondrial biogenesis with an emphasis on the PGC-1α modulator, in order to uncover the new potential pharmacological targets and ameliorate the mitochondrial disease.
2 Materials and methods
2.1 Cell lines and cell culture
Human kidney epithelial 293T cells were supplied by the Biochemistry and Molecular Biology Laboratory of Yunnan University, China. Mouse embryonic fibroblasts C3H/10T1/2 clone 8 (CH1) were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences, China. Cells were cultured with Dulbecco’s minimum essential medium (DMEM; Hyclone, Logan, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Hyclone, Logan, USA) and 1% (v/v) penicillin/streptomycin (Hyclone, Logan, USA) at 37 °C in 5% CO2.
2.2 Isolation of human PGC-1α gene
Total RNA was extracted from human epithelial 293T cells using the TRIzol reagent (Life Technologies, Foster City, USA). Complementary DNA (cDNA) was synthesized using the SuperScript First-Strand Kit (TaKaRa, Dalian, China). Primers for reverse transcription-polymerase chain reaction (RT-PCR) were designed by Primer Premier 5.0 software to amplify the complete PGC-1α open reading frame (ORF) specified in the human PGC-1α gene reference sequence at GenBank (accession No. NM_013261.3). Primer sequences were as follows: PGC-1α-BamHI-F: 5′-GGA TCC ATG GCG TGG GAC ATG TGC AAC-3′; PGC-1α-SalI-R: 5′-GTC GAC TTA CCT GCG CAA GCT TCT CTG AG-3′. BamHI and SalI restriction enzyme sites are underlined, respectively. For PCR, the reaction mixture contained 20 ng/µl cDNA template, MgCl2, primers, 10 mmol/L dNTP, 10× reaction buffer, Taq DNA polymerase, and double distilled water (ddH2O). The PCR reaction was conducted for 30 cycles, with each cycle consisting of steps at 94 °C for 2 min, 94 °C for 40 s, 66 °C for 30 s, and 72 °C for 150 s. After 30 cycles, samples were subjected to a final step at 72 °C for 10 min. PCR products were separated by agarose gel electrophoresis and the PGC-1α cDNA fragment was purified using a DNA recovery kit (TaKaRa, Dalian, China).
2.3 Construction of PGC-1α recombinant plasmid
To construct pMD18T-PGC-1α, we prepared a reaction mixture containing pMD18T vector, PGC-1α DNA, and T-Vector Solution I. Samples were incubated at 16 °C overnight, and positive clones were sent to Shanghai Sangon Co. for DNA sequencing. After sequence confirmation, pMD18T-PGC-1α and pBABEneo (Addgene, USA) were digested by restriction enzymes BamHI and SalI (TaKaRa, Dalian, China) to construct pBABEneo-PGC-1α. The digested products were purified by a DNA recovery kit and ligated. The ligation mixture was transformed into DH5α competent cells (TransGen, Beijing, China). PCR was used to screen for positive clones and the recombinant plasmid of pBABEneo-PGC-1α was sequenced by Shanghai Sangon Co., China.
2.4 Virus packaging and cellular stable transfection
Recombinant pBABEneo-PGC-1α vector, packaging plasmid pCLECO, and X-tremeGENE HP DNA Transfection Reagent (Roche, Basel, Switzerland) were added into 10% FBS DMEM medium, mixed gently, and incubated at room temperature for 20 min. The mixture was added dropwise into 293T cells in a 10-cm plate. After 48 h, the supernatant containing pBABEneo-PGC-1α virus was collected and filtered using a 0.45-µm filter. CH1 cells were then infected with the virus. After 48 h of culture, aminoglycoside antibiotic G418 (Gibco-BRL, Carlsbad, USA) was added into the medium at a final concentration of 600 µg/ml to select CH1 cells with stable expression of PGC-1α. PGC-1α expression was confirmed by Western blotting (Chen et al., 2014).
2.5 RNA interference of PGC-1α
Three double strands of small interference RNAs (siRNAs; 1009, 1523, and 853) fragments of PGC-1α were synthesized by GenePharma Co. (Shanghai, China). These siRNAs were designed according to the mRNA (NM_013261.3) loci of 1009 (target: 5′-CAC CAC TCC TCC TCA TAA A-3′), 1523 (target: 5′-GGA CAG TGA TTT CAG TAA T-3′), and 853 (5′-GTC GCA GTC ACA ACA CTT A-3′). These siRNA sequences are as follows: 1009, sense 5′-CAC CAC UCC UCC UCA UAA ATT-3′ and antisense 5′-UUU AUG AGG AGG AGU GGU GTT-3′; 1523, sense 5′-GGA CAG UGA UUU CAG UAA UTT-3′ and antisense 5′-AUU ACU GAA AUC ACU GUC CTT-3′; 853, sense 5′-GUC GCA GUC ACA ACA CUU ATT-3′ and antisense 5′-UAA GUG UUG UGA CUG CGA CTT-3′. CH1 cells with PGC-1α stable expression (CH1 PGC-1α) were seeded and cultured until they reached 60% confluence. A mixture containing the PGC-1α interference fragment and a green fluorescent control for measuring transfection efficiency (FAM fragment) was transfected into CH1 PGC-1α cells using X-tremeGENE HP DNA transfection reagent. Transfection efficiency was measured after 48 h.
2.6 Method of treatment by oligomycin A or hydrogen peroxide
When cells reached 70% confluence, they were incubated with oligomycin A (Sigma, St. Louis, USA), an inhibitor of ATP synthase, or hydrogen peroxide (Sangon, Shanghai, China) for varying lengths of time to find the optimal treatment. Oligomycin A was dissolved in ethanol at a concentration of 1 mg/ml and cells were treated for 6, 12, 24, 36, and 48 h. The time point of 24 h was chosen as the optimal time point for reduction of the cellular ATP level. Cells were treated with 100 or 200 µmol/L of hydrogen peroxide for 5 h, a treatment course in which cellular hydrogen peroxide content was high but cell death was not induced. Effects of the treatment on protein expression and cell growth were determined using Western blotting and flow cytometry.
2.7 Western blot
Cells were collected and washed twice with phosphate buffer solution (PBS), after which they were incubated in an ice-cold RIPA lysis buffer (Beyotime, Jiangsu, China) on ice for 20 min and centrifuged at 10 000g for 15 min at 4 °C. The supernatant was collected and the protein concentration was quantified with a BCA kit (Dingguo, Beijing, China). Whole cell lysate (50 µg) was loaded onto a 12.5% polyacrylamide gel for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a polyvinylidene fluoride (PVDF) membrane with 100 V constant voltage for 2 h. The membrane was then blocked with 5% (0.05 g/ml) milk and incubated with primary antibody (at 1:1000 (v/v) dilution) at 4 °C overnight. After being washed 3 times with PBST (PBS solution contain 0.1% Tween 20), the membrane was incubated with a secondary antibody (at 1:2000 (v/v) dilution) at room temperature for 2 h, and protein was detected with an enhanced chemiluminescence kit (Thermo scientific, Boston, USA). PGC-1α, CyclinD1, and CyclinB1 antibodies were purchased from Cell Signaling Technology Co. (CST, Boston, USA), and an antitubulin antibody was obtained from Beyotime Co. (Jiangsu, China) (Chen et al., 2014). To quantify relative protein expression, Western blots were analyzed using ImageJ software.
2.8 Cell cycle analysis by flow cytometry
Cells were harvested and washed in cold PBS, then incubated in 50 µg/ml of propidium iodide (PI) solution (containing 0.03% TritonX-100) at room temperature for 20 min. For each sample, at least 2×105 cells/ml were analyzed by a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, USA). Cell cycle profiles were calculated by Modfit LT software.
2.9 Cell viability
Cells were seeded into 96-well plates. Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay every 12 h during a 60-h period. Cell Titer 96 Aqueous One Solution (Promega, Madison, USA) was added to each well and cells were incubated at 37 °C for 2 h. The optical density at 490 nm (OD490) was read using a microplate reader (Bio-Rad, Tokyo, Japan).
2.10 ROS measurement
Cells were collected and resuspended in PBS at 1×105 cells/ml. MitoSOX Red dye (5 µmol/L; mitochondrial ROS specific, Invitrogen-Life Technologies, New York, USA) was added to the cells, after which they were incubated at 37 °C for 30 min. Untreated cells served as negative controls. Red fluorescence was detected using a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, USA) (Hahm et al., 2014; Xu et al., 2015).
2.11 Comparative assay of ATP
Cells were collected and resuspended in PBS at 1×106 cells/ml. ATP lysis buffer was added into cells and incubated on ice for 30 min. The cell lysate was centrifuged at 12 000 r/min for 10 min, after which ATP levels in the supernatant were measured using an ATP assay kit (Beyotime, Jiangsu, China). Bioluminescence was measured on a Pi-102 fluorescence luminometer (Hygiena, New York, USA). Cellular ATP content (nmol/L) was calculated using an ATP standard and normalized using a BCA protein assay kit (Xiong et al., 2012).
2.12 Statistical analysis
Statistical analysis was performed using the SPSS version 16.0 software. All experiments were repeated at least three times and the evaluation data parameters are reported as mean±standard deviation (SD).
3 Results
3.1 pBABE-PGC-1α plasmid construction
Retroviral vector pBABEneo was used to construct a pBABE-PGC-1α recombinant plasmid containing the PGC-1α-coding sequence. PCR was used to confirm the presence of the PGC-1α-coding sequence, which had an expected PCR product size of 2396 bp (Fig. 1a). We then sub-cloned PGC-1α cDNA into a pMD18T plasmid containing BamHI and SalI restriction sites to construct pMD18T-PGC-1α. Positive clones were sequenced using pMD18T primers and found to have the correct PGC-1α cDNA sequence. Then pMD18T-PGC-1α (Fig. 1b, Lane 1) and pBABEneo were digested with BamHI and SalI. Agarose gel electrophoresis showed that the correct product size was obtained after digestion with the two enzymes (Fig. 1b, Lane 2). The digested PGC-1α fragment and pBABEneo were purified using a DNA recovery kit and ligated with T4 ligase. The ligation product of pBABEneo-PGC-1α (about 7700 bp in size; Fig. 1c, Lane 1) was confirmed by sequencing and transformed into DH5α competent cells. Positive bacterial clones were digested and showed the correct product size (Fig. 1c, Lane 2). These results indicated successful construction of the pBABE-PGC-1α recombinant plasmid.
Strategy for construction of pBABEneo-PGC-1 α expression vector
(a) Agarose gel analysis of PGC-1α cDNA PCR product. M: 1 kb DNA ladder; Lanes 1 and 2: PGC-1α cDNA. (b) Gel image of pMD18T-PGC-1α digested with BamHI and SalI. Lane 1: pMD18T-PGC-1α plasmid; Lane 2: after digestion. (c) Gel image of pBABEneo-PGC-1α digested by BamHI and SalI. Lane 1: pBABEneo-PGC-1α plasmid; Lane 2: after digestion
3.2 Establishment of cell lines overexpressing PGC-1α or with PGC-1α knockdown
To make virus particles, 293T cells were co-transfected with pBABEneo-PGC-1α recombinant plasmid and packaging plasmid pCLECO using X-tremeGENE HP DNA transfection reagent. As a control, cells were transfected with the empty vector pBABEneo. Cells were harvested after 48 h and PGC-1α expression was measured using Western blotting. We found that PGC-1α was highly expressed in pBABEneo-PGC-1α co-transfected cells but not in cells transfected with the empty control vector. Supernatant containing pBABEneo-PGC-1α virus or control virus was collected to infect the host CH1 cells. After infection, CH1 cells with stable expression of PGC-1α or control vector were selected in culture media supplemented with the antibiotic G418. As expected, PGC-1α was expressed at higher levels in pBABEneo-PGC-1α stable CH1 cells (PGC-1α), compared with cells containing the pBABEneo empty vector (PB) (Fig. 2a). To study the function of PGC-1α, we transfected cells overexpressing PGC-1α with siRNA (Si) to knock down PGC-1α. In this process, the efficiency of siRNA had been demonstrated in PGC-1α cells by NC (non-targeting control) and FAM (siRNA has fluorescence) (Fig. 2b). To study the function of PGC-1α, we tested the efficiency of three siRNAs (1009, 1523, and 853) in PB (Figs. 2c and 2d) and PGC-1α (Figs. 2e and 2f) cell lines through the PGC-1α expression level. The level of PGC-1α expression showed no significant difference between PB and Si groups. However, there is a significant difference between PGC-1α and Si groups, which could be due to the much lower endogenic PGC-1α expression in PB cells. The effect of 853 was the best among these three siRNAs (Figs. 2e and 2f), so we chose 853 for subsequent experiments. These results confirmed the cell lines in which various levels of PGC-1α established, and which in subsequent experiments were used to determine the role of PGC-1α in mitochondrial function and cell proliferation.
Establishment of a stable CH1 cell line expressing PGC-1 α and RNA interference
(a) Expression of PGC-1α protein in PB (pBABEneo empty vector) and PGC-1α (pBABEneo-PGC-1α) cell lines; (b) The efficiency of small interference RNA (siRNA) was detected by flow cytometry (NC as non-targeting control and FAM as efficiency detection with fluorescence); (c) Expression of PGC-1α protein in PB and PB cells that were interfered by three siRNA (1009, 1523, and 853); (d) Semi-quantification of PGC-1α protein expression in (c); (e) Expression of PGC-1α protein in PB, PGC-1α, and PGC-1α cells that were interfered by three siRNA (1009, 1523, and 853); (f) Semi-quantification of PGC-1α protein expression in (e). ** P<0.01, * P<0.05. Data are expressed as mean±SD (n=3)
3.3 Mitochondrial function and cell proliferation in CH1 PGC-1α cells
Cellular ROS and ATP contents can be used as markers of proper mitochondrial function. As PGC-1α is an important regulator of mitochondrial function, we sought to determine whether PGC-1α expression level affects ROS and ATP contents. Using the fluorescent superoxide indicator MitoSOX Red, we found that ROS levels in PGC-1α cells were lower than those in PB and Si cells (Fig. 3a). In contrast, ATP assay showed that ATP levels in PGC-1α cells were higher than those in PB and Si cells (P<0.01; Fig. 3b). These results showed that PGC-1α reduced ROS and increased ATP levels in a concentration-dependent manner. To determine the role of PGC-1α in cell growth, cell proliferation rates were determined using an MTS kit. PGC-1α cells proliferated significantly faster than PB cells and Si cells during the time course examined (Fig. 3c). These results suggest that CH1 PGC-1α cells can be used as a model for studying the underlying mechanisms by which PGC-1α regulates cell growth.
3.4 Comparative analysis of cell cycle profile
In order to check whether PGC-1α functions on cell growth and cell proliferation, the cell cycle profiles of PGC-1α, PB, and Si cell lines were measured by flow cytometry. We observed greater S phase accumulation in PGC-1α cells ((53.50±2.35)%) than in PB cells ((48.14±1.32)%) or Si cells ((47.32±1.27)%) (Figs. 4a and 4b). There was just a slight difference in G2/M progression between PGC-1α and Si groups. The CyclinD1 and CyclinB1 play important roles in cell cycle regulation through their interactions with CDKs. Specifically, CyclinD1 regulates the transition from G1 to S phase, whereas CyclinB1 regulates the transition from G2 to M phase. We found that both CyclinD1 and CyclinB1 were overexpressed in CH1 PGC-1α cells (Figs. 4c–4e). These results were also confirmed by immunofluorescence images which are presented in supplementary information (Fig. S1). Up-regulation of CyclinD1 could contribute to the S phase accumulation. However, the up-regulated CyclinB1 did not change the proportion of G2/M phase cells. There are two possibilities: the up-regulated CyclinB1 was located in the cytoplasm instead of the nucleus, or it remained dephosphorylated. Both of them block the activity of CyclinB1. More experiments need to be conducted to determine which is the cause.
Cell cycle profiles and CylinD1/B1 expression in CH1 cells
Cell cycle profiles (a) and phase proportions (b) of PB, PGC-1α, and Si cells; (c) The expressions of CyclinD1 and CyclinB1 in PB, PGC-1α, and Si cells were detected by Western blotting; (d) Semi-quantification of CyclinB1 protein expression in (c); (e) Semi-quantification of CyclinD1 protein expression in (c); (f) Semi-quantification of PGC-1α protein expression in (c). ** P<0.01. Data are expressed as mean±SD (n=3)
3.5 Analysis of cell cycle after oligomycin A treatment
There are several possible mechanisms by which PGC-1α could promote S phase accumulation. PGC-1α may directly upregulate CyclinD1 and CyclinB1 to increase S phase accumulation and accelerate cell growth. However, another possibility is that the increased ATP or the decreased ROS induced by PGC-1α serve as signals to regulate CyclinD1 and CyclinB1. An increasing body of evidence indicates that ROS and ATP are important players in various cellular regulation pathways. As such, we sought to externally manipulate ATP and ROS levels to determine their influence on CyclinD1 and CyclinB1.
To decrease ATP generation, we blocked the mitochondrial respiratory chain by culturing cells in media supplemented with oligomycin A for 24 h. Interestingly, we found that CyclinD1 was down-regulated by oligomycin A treatment in a dose-dependent manner (Figs. 5a and 5b), whereas CyclinB1 showed no significant change (Figs. 5a and 5c). In addition, PGC-1α expression was maintained at a constant level in each treatment condition (Fig. 5d). Further analysis of the cell cycle profile showed less S phase accumulation in PGC-1α cells ((56.47±3.26)%) after oligomycin A treatment than in untreated cells ((30.94±1.78)%; Figs. 5e and 5f). These results suggest that PGC-1α regulates CyclinD1, but not CyclinB1, through its influence on ATP.
Cell cycle profiles and changes in CyclinD1/B1 levels in CH1-PGC-1a after 24 h of oligomycin A treatment
(a) Protein expressions of CyclinB1, CyclinD1, and PGC-1α. PGC-1α cells were treated with oligomycin A (0, 5, and 8 µg/ml). PB was used as a control for PGC-1α. Semi-quantification of CyclinD1 (b), CyclinB1 (c), and PGC-1α (d) in (a). (e) Profiles of cell cycles. (f) The percentage of CH1 cells in each phase. ** P<0.01, * P<0.05. Data are expressed as mean±SD (n=3)
3.6 Analysis of cell cycle after H2O2 treatment
We found that ROS levels were low in CH1 PGC-1α cells (Fig. 4a). To explore whether ROS function as a signal to influence cell cycle progression, we added exogenous H2O2 to increase ROS and detected the expression of CyclinB1 and CyclinD1. Western blotting (Fig. 6a) showed H2O2 exposure decreased CyclinB1 (Fig. 6b), but not CyclinD1 (Fig. 6c). Importantly, we observed that PGC-1α was maintained at a constant level in each treatment condition (Fig. 6d), suggesting that CyclinB1 was down-regulated by H2O2, and not by PGC-1α. Finally, cell cycle profiling showed that increased H2O2 inhibited cell growth, as (56.15±2.31)% of untreated PGC-1α cells were in S phase, compared with (31.37±1.33)% of cells treated with 100 µmol/L of H2O2 (Figs. 6e and 6f).
Cell cycle profiles and changes in CyclinD1/B1 in CH1-PGC-1 α after 5 h of H 2 O 2 exposure
(a) Probe for CyclinB1, CyclinD1, and PGC-1α. PGC-1α cells were exposed to H2O2 (0, 100, and 200 µmol/L). PB served as a control for PGC-1α. Semi-quantification of CyclinD1 (b), CyclinB1 (c), and PGC-1α (d) in (a). (e) Profile of cell cycle. (f) The percentage of CH1 cells in each phase. ** P<0.01. Data are expressed as mean±SD (n=3)
4 Discussion
A large body of experimental evidence demonstrates that mitochondrial OXPHOS significantly contributes to ATP supply and ROS production. However, fewer papers reported that association of the mitochondrial respiratory chain complexes form higher supramolecular structures, which are called mitochondrial respiratory supercomplexes. These supercomplexes can take electrons from NADH to reduce oxygen (Acín-Pérez et al., 2008; Althoff et al., 2011; Vartak et al., 2013). Mitochondria exert a pivotal role in mediating cell proliferation and apoptosis (Zhang et al., 2007), and mitochondrial dysfunction is a hallmark of many genetic diseases and tumor progression. As mitochondrial biogenesis can be manipulated to compensate for deficits in energy, it has been recently proposed as a potential therapeutic target for treatment of mitochondrial disease (Valero, 2014). Recently, several independent groups identified PGC-1α as a key modulator of mitochondrial OXPHOS to increase the level of ATP (Rohas et al., 2007; Won et al., 2010), and suggested that PGC-1α may be involved in aging and tumorigenesis. Furthermore, both mitochondrial dysfunction and down-regulation of PGC-1α are present in Huntington’s disease, Alzheimer’s disease, and Parkinson’s disease, providing further support for a relationship between mitochondrial function and PGC-1α (Miraglia et al., 2015; Rice et al., 2015). Although a mitochondrial origin for these diseases has been proposed (Falk et al., 2015), regulation of mitochondrial biogenesis in the context of these diseases has not been extensively studied. In this study, we established cell lines in which PGC-1α was overexpressed or knocked down to investigate the signaling pathways by which PGC-1α could influence cell growth.
We found that elevated PGC-1α expression increased ATP level, reduced ROS production, and accelerated cell proliferation. The increase of ATP could be due to more mitochondria content (mitochondrial content was detected with the result presented in Fig. S2). PGC-1α promotes mitochondrial biogenesis through expression of mitochondrial proteins and activation of p38/MAPK pathway (Acín-Pérez et al., 2008; Luckhart et al., 2013). These results confirmed that PGC-1α has an important role in prompting ATP production and clearing excessive ROS, and mitochondrial respiratory supercomplexes also enhance ATP and decrease ROS through structure variant, so we suggested that PGC-1α may be a positive regulator of mitochondrial respiratory supercomplex formation, but the mechanism remains unclear. A major focus of our study was to address the influence of PGC-1α in the context of mitochondrial function and cell cycle progression. We hypothesized that PGC-1α regulates cell cycle proteins and triggers cell proliferation indirectly, through its modulation of ATP and ROS levels.
To test our hypothesis, we manipulated mitochondrial function to attenuate ATP production and accumulate ROS in order to determine whether they would reduce the oncogenic properties of PGC-1α cells. Importantly, PGC-1α levels remained constant throughout treatment. The electron transport chain (ETC) is responsible for OXPHOS-mediated mitochondrial respiration (Chen et al., 2010). Our results demonstrated that oligomycin A, a classic mitochondrial complex-III inhibitor, blocked ATP production and disrupted progression to S phase. We then measured levels of the well-known G1/S transition protein CyclinD1, and the G2/M checkpoint regulatory protein CyclinB1 (Schick et al., 2007; Dalton, 2013). We found that CyclinD1, but not CyclinB1, was down-regulated, while PGC-1α was maintained at a constant level. This result suggests that ATP can regulate cell cycle progression through CyclinD1, and may do so as a consequence of its up-regulation by PGC-1α. We also detected the expression levels of CyclinB1 and CyclinD1 by using antimycin A-treated PGC-1α cells. The result showed that CyclinB1 and CyclinD1 were down-regulated (Fig. S3).
We also found a relationship between G2/M phase restriction and CyclinB1 up-regulation in CH1 PGC-1α cells. These cells had lower ROS levels than cells in which PGC-1α was not upregulated. ROS have been identified as important mediators of cell proliferation, cell death, and mitochondrial membrane depolarization (Wood et al., 2003; Marinho et al., 2014). We attempted to find whether H2O2 enhanced oxidative burst, the transient production of large amounts of ROS, plays a role in controlling cell cycle checkpoints and in inhibition of cell proliferation. Upon treatment with H2O2, we found that cell proliferation was inhibited and CyclinB1 was down-regulated, but CyclinD1 was unaffected.
In summary, our results suggest that mitochondrial complexIII is a target of PGC-1α. Inhibition of ATP production restricted the G1/S phase transition. In contrast, accumulation of ROS involved G2/M phase arrest through down-regulation of CyclinB1. Taken together, these findings suggest that controlling PGC-1α expression level could represent a reliable and efficient way to maintain ATP and ROS within levels compatible with signaling, while ensuring a robust and reliable energy supply. As such, it holds great potential for treatment of mitochondrial diseases.
Compliance with ethics guidelines
Xu-feng FU, Kun YAO, Xing DU, Yan LI, Xiu-yu YANG, Min YU, Mei-zhang LI, and Qing-hua CUI declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
List of electronic supplementary materials
Fig. S1 Immunofluorescence picture of CyclinD1 and CyclinB1 in PB, PGC-1α, and Si cells
Fig. S2 Mitochondrial content indicated by MitoTracker Green fluorescence (analyzed by flow cytometry)
Fig. S3 Change of CyclinD1/B1 levels in CH1-PGC-1α after 24 h of antimycin A treatment
References
Acín-Pérez, P.R., Fernández, S.P., Peleato, M.L., et al., 2008. Respiratory active mitochondrial supercomplexes. Mol. Cel., 32(4):529–539. http://dx.doi.org/10.1016/j.molcel.2008.10.021
Althoff, T., Mills, D.J., Popot, J.L., et al., 2011. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J., 30(22): 4652–4664. http://dx.doi.org/10.1038/emboj.2011.324
Bertoli, C., Skotheim, J.M., Bruin, R.A., 2013. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol., 14(8):518–528. http://dx.doi.org/10.1038/nrm3629
Cannon, B., Houstek, J., Nedergaard, J., 1998. Brown adipose tissue: more than an effector of thermogenesis? Ann. NY Acad. Sci., 856:171–187. http://dx.doi.org/10.1111/j.1749-6632.1998.tb08325.x
Chaturvedi, R.K., Beal, M.F., 2013. Mitochondrial diseases of the brain. Free Radical Biol. Med., 63:1–29. http://dx.doi.org/10.1016/j.freeradbiomed.2013.03.018
Chen, G., Dai, J., Tan, S., et al., 2014. MTERF1 regulates the oxidative phosphorylation activity and cell proliferation in HeLa cells. Acta Biochim. Biophys. Sin., 46(6):512–521. http://dx.doi.org/10.1093/abbs/gmu029
Chen, Q., Yin, G., Stewart, S., et al., 2010. Isolating the segment of the mitochondrial electron transport chain responsible for mitochondrial damage during cardiac ischemia. Biochem. Biophys. Res. Commun., 397(4):656–660. http://dx.doi.org/10.1016/j.bbrc.2010.05.137
Dalton, S., 2013. G1 compartmentalization and cell fate coordination. Cel., 155(1):13–14. http://dx.doi.org/10.1016/j.cell.2013.09.015
Falk, M.J., Shen, L., Gonzalez, M., et al., 2015. Mitochondrial Disease Sequence Data Resource (MSeqDR): a global grass-roots consortium to facilitate deposition, curation, annotation, and integrated analysis of genomic data for the mitochondrial disease clinical and research communities. Mol. Genet. Metab., 114(3):388–396. http://dx.doi.org/10.1016/j.ymgme.2014.11.016
Hahm, E.R., Sakao, K., Singh, S.V., 2014. Honokiol activates reactive oxygen species-mediated cytoprotective autophagy in human prostate cancer cells. Prostat., 74(12): 1209–1221. http://dx.doi.org/10.1002/pros.22837
Lehman, J.J., Barger, P.M., Kovacs, A., et al., 2000. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest., 106(7): 847–856. http://dx.doi.org/10.1172/JCI10268
Lin, J., Handschin, C., Spiegelman, B.M., 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab., 1(6):361–370. http://dx.doi.org/10.1016/j.cmet.2005.05.004
Löbrich, M., Jeggo, P.A., 2007. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cance., 7(11):861–869. http://dx.doi.org/10.1038/nrc2248
Luckhart, S., Giulivi, C., Drexler, A.L., et al., 2013. Sustained activation of Akt elicits mitochondrial dysfunction to block Plasmodium falciparum infection in the mosquito host. PLoS Pathog., 9(2):e1003180. http://dx.doi.org/10.1371/journal.ppat.1003180
Malumbres, M., Barbacid, M., 2009. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cance., 9(3):153–166. http://dx.doi.org/10.1038/nrc2602
Marinho, H.S., Real, C., Cyrne, L., et al., 2014. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol., 2:535–562. http://dx.doi.org/10.1016/j.redox.2014.02.006
McBride, H.M., Neuspiel, M., Wasiak, S., 2006. Mitochondria: more than just a powerhouse. Curr. Biol., 16(14): R551–R560. http://dx.doi.org/10.1016/j.cub.2006.06.054
Meirhaeghe, A., Crowley, V., Lenaghan, C., et al., 2003. Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-γ co-activator 1β) gene in vitro and in vivo. Biochem. J., 373(Pt 1): 155–165. http://dx.doi.org/10.1042/bj20030200
Miraglia, F., Betti, L., Palego, L., et al., 2015. Parkinson’s disease and a-synucleinopathies: from arising pathways to therapeutic challenge. Cent. Nerv. Syst. Agents Med. Chem., 15(2):109–116. http://dx.doi.org/10.2174/1871524915666150421114338
Pieczenik, S.R., Neustadt, J., 2007. Mitochondrial dysfunction and molecular pathways of disease. Exp. Mol. Pathol., 83(1):84–92. http://dx.doi.org/10.1016/j.yexmp.2006.09.008
Rice, A.C., Ladd, A.C., Bennett, J.P., 2015. Postmortem Alzheimer’s disease hippocampi show oxidative phosphorylation gene expression opposite that of isolated pyramidal neurons. J. Alzheimer’s Dis., 45(4):1051–1059. http://dx.doi.org/10.3233/JAD-142937
Rohas, L.M., St-Pierre, J., Uldry, M., et al., 2007. A fundamental system of cellular energy homeostasis regulated by PGC-1a. PNA., 104(19):7933–7938. http://dx.doi.org/10.1073/pnas.0702683104
Schick, V., Majores, M., Fassunke, J., et al., 2007. Mutational and expression analysis of CDK1, cyclinA2 and cyclinB1 in epilepsy-associated glioneuronal lesions. Neuropathol. Appl. Neurobiol., 33(2):152–162. http://dx.doi.org/10.1111/j.1365-2990.2006.00788.x
Shiota, T., Traven, A., Lithgow, T., 2015. Mitochondrial biogenesis: cell-cycle-dependent investment in making mitochondria. Curr. Biol., 25(2):R78–R80. http://dx.doi.org/10.1016/j.cub.2014.12.006
Valero, T., 2014. Mitochondrial biogenesis: pharmacological approaches. Curr. Pharm. Des., 20(35):5507–5509. http://dx.doi.org/10.2174/138161282035140911142118
Vartak, R., Porras, C.A., Bai, Y., 2013. Respiratory supercomplexes: structure, function and assembly. Protein Cel., 4(8):582–590. http://dx.doi.org/10.1007/s13238-013-3032-y
Vega, R.B., Huss, J.M., Kelly, D.P., 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor a in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol., 20(5):1868–1876. http://dx.doi.org/10.1128/MCB.20.5.1868-1876.2000
Wallace, D.C., 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet., 39(1):359–407. http://dx.doi.org/10.1146/annurev.genet.39.110304.095751
Weinberg, F., Chandel, N.S., 2009. Mitochondrial metabolism and cancer. Ann. NY Acad. Sci., 1177(1):66–73. http://dx.doi.org/10.1111/j.1749-6632.2009.05039.x
Won, J.C., Park, J.Y., Kim, Y.M., et al., 2010. Peroxisome proliferator-activated receptor-γ coactivator 1-a overexpression prevents endothelial apoptosis by increasing ATP/ADP translocase activity. Arterioscler. Thromb. Vasc. Biol., 30(2):290–297. http://dx.doi.org/10.1161/ATVBAHA.109.198721
Wood, Z.A., Poole, L.B., Karplus, P.A., 2003. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Scienc., 300(5619):650–653. http://dx.doi.org/10.1126/science.1080405
Wu, Z., Puigserver, P., Andersson, U., et al., 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cel., 98(1):115–124. http://dx.doi.org/10.1016/S0092-8674(00)80611-x
Xiong, W., Jiao, Y., Huang, W., et al., 2012. Regulation of the cell cycle via mitochondrial gene expression and energy metabolism in HeLa cells. Acta Biochim. Bioph. Sin., 44(4):347–358. http://dx.doi.org/10.1093/abbs/gms006
Xu, H., Lyu, S., Xu, J., et al., 2015. Effect of lipopolysaccharide on the hemocyte apoptosis of Eriocheir sinensis. J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol)., 16(12): 971–979. http://dx.doi.org/10.1631/jzus.B1500098
Zhang, Y., Ba, Y., Liu, C., et al., 2007. PGC-1α induces apoptosis in human epithelial ovarian cancer cells through a PPARγ-dependent pathway. Cell Res., 17(4): 363–373. http://dx.doi.org/10.1038/cr.2007.11
Acknowledgements
We would like to thank all the members of our laboratory for their participation in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
The two authors contributed equally to this work
Project supported by the National Natural Science Foundation of China (Nos. 31160237 and 81360310) and the Graduate Student Research Innovation Project of Yunnan University (No. YNUY201455), China
ORCID: Xu-feng FU, http://orcid.org/0000-0002-4101-1000
Supplemental Information
Rights and permissions
About this article
Cite this article
Fu, Xf., Yao, K., Du, X. et al. PGC-1α regulates the cell cycle through ATP and ROS in CH1 cells. J. Zhejiang Univ. Sci. B 17, 136–146 (2016). https://doi.org/10.1631/jzus.B1500158
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1500158
Keywords
- Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α)
- Mitochondria
- Oxidative phosphorylation (OXPHOS)
- Cell cycle
- CyclinD1
- CyclinB1