Abstract
Eukaryotic gene expression is controlled by different levels of biological events, such as transcription factors regulating the timing and strength of transcripts production, alteration of transcription rate by RNA processing, and mRNA stability during RNA processing and translation. RNAs, especially mRNAs, are relatively vulnerable molecules in living cells for ribonucleases (RNases). The maintenance of quality and quantity of transcripts is a key issue for many biological processes. Extensive studies draw the conclusion that the stability of RNAs is dedicated-regulated, occurring co- and post-transcriptionally, and translation-coupled as well, either in the nucleus or cytoplasm. Recently, RNA stability in the nucleus has aroused much research interest, especially the stability of newly-made transcripts. In this article, we summarize recent progresses on mRNA stability in the nucleus, especially focusing on quality control of newly-made RNA by RNA polymerase II in eukaryotes.
Similar content being viewed by others
1 Introduction
Gene expression is a fundamental and essential event for living cells in every organism, which is controlled by sophisticate networks of many processes and pathways, involving RNA transcription, post-transcriptional RNA processing and modification, RNA export, translation, and messenger RNA (mRNA) stability. mRNA levels in living cells represent the balance between production (transcription) and decay (mRNA degradation). mRNA stability, as an important factor in the control of gene expression, only depends on degradation rates of mRNA and does not relate to the steady-state levels of mRNA. In living cells, mRNAs show a variety of turnovers and degradation rates (Wang et al., 2002; Yang et al., 2003; Grigull et al., 2004).
mRNA turnover in bacteria is believed to be initiated by an endonucleolytic cleavage, which is followed by a 3’-5’ decay. Degradation of mRNAs is mainly executed by relatively conserved degradosome, though this consists of many ribonucleases which vary in different species (Babitzke and Kushner, 1991; Taraseviciene et al., 1991; Even et al., 2005; Mathy et al., 2007; Shahbabian et al., 2009). In contrast to eukaryotic cells, mRNA degradation in bacteria is presumed to be more tightly linked to both transcription and translation, due to the lack of spatial separation of chromatin and translation machinery by nuclear membrane. However, Montero Llopis et al. (2010) suggest that mRNA turnover in bacteria shares similar characteristics with that in eukaryotes, i.e., degradation of mRNA is a spatial event, happening at the nuclear region and being associated with chromatin.
In eukaryotes, many mRNA degradation pathways in the cytoplasm have been extensively studied. Almost all of them require the same major degradation machineries, either XRN1, the main exoribonuclease responsible for 5’-3’ degradation, or exosome for 3’-5’ degradation, regardless of normal or aberrant mRNAs. Many proposed mRNA degradation mechanisms take place in the cytoplasm, like microRNA mediated mRNA degradation and AU-rich element mediated mRNA decay (AMD), and some mRNA decays due to the occurrence of aberrant translations, such as Staufen 1-mediated mRNA decay (SMD), nonsense-mediated mRNA decay (NMD), no-go decay (NGD), and non-stop decay (NSD) (Peng et al., 1998; van Hoof et al., 2002; Kim et al., 2005; Doma and Parker, 2006; Brogna and Wen, 2009; Chekulaeva and Filipowicz, 2009).
Although cytoplasmic mRNA decay seems to be the dominant mRNA turnover in eukaryotes, we believe that nuclear RNA degradation and recycle are far more ubiquitous than we expected. It is clear that typically, among RNAs produced by RNA polymerase II (RNP II), less than 10% of nucleotides in human cells are kept in mature RNAs because more than 90% nucleotides are spliced out as introns and recycled in the nucleus. Until recently, we still did not know how many RNP II transcripts are lost in the nucleus by degradation. For example, only 50% transcripts of the longest gene in the human genome, Dystrophin, are managed to be produced as a mature nuclear mRNA (Jackson et al., 2000). Jackson et al. (2000) also summarized that only a minor proportion, about 30% of transcripts, is processed to be mRNA and exported to the cytoplasm. The predominant population of nuclear transcripts in human cells seems to be unpolyadenylated and poorly spliced, which renders these transcripts unable to be exported. Hence, they are restrained at the transcription site or the proximity of the nuclear pore, and eventually degraded in the nucleus (Jackson et al., 2000). Moreover, recent studies focusing on exosome targets in transcriptome revealed that massive amounts of RNA precursors are degraded before they enter functional pathways in Saccharomyces cerevisiae (Gudipati et al., 2012; Schneider et al., 2012). All of these observations announced the presence of extensive nuclear RNA degradation in eukaryotic cells.
2 Protein/protein complex for RNA degradation in nucleus
2.1 Exosome
As mentioned above, the variety of transcription processes generates a huge amount of ‘waste nuclear RNA’, which requires many ribonucleolytic activities to be recycled. One dominant protein complex implicated in nuclear RNA turnover is exosome. Exosome is a conserved ~400-kD hetero-multimeric protein complex in eukaryotes, containing nine core components (named as Exo9) and acting as 3’-5’ exoribonuclease and endoribonuclease in association with some cofactors (Mitchell et al., 1997; Hilleren et al., 2001; Houseley et al., 2006; Vanacova and Stefl, 2007). In eukaryotes, there are two general forms of exosomes: one is the cytoplasmic exosome that contains the nine-subunit core (Exo9) plus Rrp44p (named as Exo10), responsible for 3’-5’ exoribonuclease and endoribonuclease activities; the other is the nuclear exosome that consists of Exo9 with Rrp44p and Rrp6p (named as Exo11) (Chen et al., 2001; Liu et al., 2006; Dziembowski et al., 2007; Tomecki et al., 2010; Malecki et al., 2013). Actually, in human cells, there are three Rrp44 homologs: DIS3 (named from yeast ‘disjunction abnormal’) that only shows exoribonuclease activity and predominantly locates in the nucleus; DIS3L that shows exo- and endoribo-nuclease activities in the cytoplasm-like yeast Rrp44p; and, DIS3L2 that does not interact with Exo9 but is involved in mRNA degradation in the cytoplasm (Tomecki et al., 2010; Malecki et al., 2013). Tomecki et al. (2010) also found a new type of exosome which is present in the nucleolar region of human cells and only contains Exo9 plus Rrp6, but its function is yet to be elucidated.
In the cytoplasm, Exo10 associates with the SKI complex that functions to unfold RNAs, and consists of Ski2p, Ski3p, and Ski8p to participate in 3’-5’ RNA degradation (Halbach et al., 2013). The nuclear exosome, Exo11, interacts with many cofactors and then functions in RNA processing and nuclear RNA degradation. Exo11 associates with Rrp47p via Rrp6p, removes the 3’-extended form of 5.8S ribosomal RNA (rRNA) precursor during rRNA biogenesis, and is also involved in the production of small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs) (Mitchell et al., 2003). It also interacts with the TRAMP complex, which consists of poly(A) polymerases Trf4/Trf5p, RNA helicase Mtr4 and the zinc knuckle proteins Air1/Air2, to remove aberrant RNAs (LaCava et al., 2005). The NNS complex (Nrd1p, Nab3p, and Sen1p) requires Exo11 to process some snRNAs and snoRNAs, which are transcribed by RNP II (Kim et al., 2006). Additionally, in human cells, Exo11 associates with the NEXT complex (nuclear exosome targeting complex), which consists of hMTR4, zinc knuckle protein ZCCHC8, and RNA binding protein RBM7, to facilitate the degradation of transcripts upstream of promoters (Lubas et al., 2011).
2.2 Rat1p/XRN2
In S. cerevisiae, Rat1p (named as XRN2 in human cells) rather than Xrn1p functions as the main exoribonuclease in 5’-3’ degradation of transcripts in the nucleus. Rat1p was initially identified in S. cerevisiae and then found to be conserved in all eukaryotes (Amberg et al., 1992; Shobuike et al., 1995; 2001; Yoo et al., 2000; West et al., 2004; Li et al., 2006). It functions essentially in RNP II transcription termination to remove the long aberrant mRNAs which associate with a chromosome and might cause deleterious effects in cells (Proudfoot, 2011). It is required after cleavage and polyadenylation of nascent transcripts to degrade the downstream cleaved RNA from 5’-3’ direction (Richard and Manley, 2009). Rat1p is also involved in the process of rRNA maturation: ITS1, one of the internal transcribed spacers (ITS1 and ITS2), locates at the flanking site of 5.8S rRNA, which is trimmed by Rat1p from the 5’ end of the rRNA precursor after endoribonuclease cleavage (Henry et al., 1994; Geerlings et al., 2000). Rat1p recently has been suggested to play an important role in telomere maintenance by degrading telomeric repeat-containing RNA (TERRA), which is a long non-coding RNA that represses telomerase activity, and thus activates telomere elongation (Luke et al., 2008). Further recent studies in human cells suggest that XRN2 degrades nascent transcripts when RNP II pauses near promoters. Brannan et al. (2012) indicate that early recruitment of termination factors XRN2 and TTF2, as well as decapping factors Edc3, Dcp1a, and Dcp2, control the bi-directional RNP II transcription by a ‘torpedo’ mechanism. In another case, XRN2 and another termination factor SetX cause RNP II pausing and premature termination at the human immunodeficiency virus (HIV) promoter, which is followed by human RRP6 processing of the HIV transcript into a small RNA to silence the RNA-dependent transcription at the HIV-1 promoter (Wagschal et al., 2012).
Rat1p forms a stable complex with Rai1p that stimulates its exoribonuclease activity (Stevens and Poole, 1995; Xue et al., 2000; Xiang et al., 2009). The complex associates with a C-terminal domain binding protein, Rrt103p, and then contributes to RNP II termination in yeast. Depletion of Rat1p or Rai1p causes defective termination of RNP II (Kim et al., 2004). Although Rai1p has homologs in human cells and Drosophila, no such interaction between XRN2 and RAI1/Rail has been detected in these cells. Interestingly, a recent study suggests that Rai1p is a pyrophosphate-hydrolase that removes the 5’ end of non-canonical capped mRNA, which is an aberrant cap lack of methylation (Jiao et al., 2010).
3 Induction of nuclear mRNA degradation by non-canonical mRNP
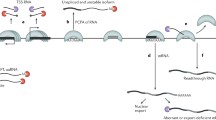
To date, we still do not have enough data to elucidate the mechanisms of nuclear mRNA decay in animal cells. Most of the studies on mRNA degradation in the nucleus use budding yeast as model organism. These studies in S. cerevisiae suggest that degradation of mRNA, by several quality control pathways, ubiquitously happens in the nucleus at many steps of mRNA production if aberrancy occurred, i.e., mRNA retention at the transcription site during transcription initiation and elongation, transcribed messenger ribonucleoprotein particle (mRNP) slowly released during termination, failure/aberration of splicing, and mRNA retention at the nuclear pore (Fig. 1).
3.1 Aberrant 5’ cap mRNP
A cap structure at the 5’ end of mRNA, one of the hallmark features of eukaryotic mRNAs, is required for the protection of transcripts from the exoribonuclease such as Rat1p that causes 5’-3’ degradation (Zhai and Xiang, 2014). Also, the mature cap associates with cap binding proteins (Cbc1p and Cbc2p), which are required for efficient co-transcriptional processing, splicing, mRNA export and translation (Gonatopoulos-Pournatzis and Cowling, 2014). The capping machinery, which directly couples to RNP II, ensures that transcripts keep to normal steady-state levels with proper transcriptions. Exceptionally, Abd1p, a methyltransferase, functions in early transcription elongation which is independent of its capping activity (Schroeder et al., 2004). It means that proteins involved in capping might have alternative activities in RNA processing and the quality control pathway. This is supported by an early study in yeast, which suggests that capping at the 5’ end mRNA is important for mRNA stability in the nucleus (Schwer et al., 1998). Conditional dysfunction (temperature-permissive mutant) of Ceg1p, the nuclear guanylyltransferase, shows low levels of some transcripts (Schwer et al., 1998). It is striking to find that the growth defect of yeast cells with mutations of Rat7p (a nuclear pore component), which usually induce nuclear mRNA retention and cause mRNA decay, was suppressed by deletion of the CBC1 gene. In these studies, in nuclear pore component mutants, several mRNAs’ fast degradations were partially restored in CBC1 mutants (Das et al., 2000; 2003). Therefore, Cbc1p likely participates in degradation of these mRNAs in export-defective strains or it may happen in the wild-type strain as well, but the mechanism is yet to be elucidated (Kuai et al., 2005). Another example has been described in Section 2.2: an aberrant cap structure induces the mRNA degradation by Rai1p under nutritional stress conditions (Jiao et al., 2010). All of these suggest that the 5’ end formation of mRNP clearly correlates to the quality control pathway in the nucleus, though it still remains mysterious.
3.2 Nuclear mRNA retention caused by pre-mRNP and aberrant mRNP
Legrain and Rosbash (1989) found that in yeast, an intron that failed to be spliced but was recognized at an early stage by spliceosome seems to be required for RNA retention in the nucleus. Later on it has been observed that pre-mRNAs/unspliced RNAs are retained in the nucleus, which is mediated by some splicing factors (Rutz and Seraphin, 2000; Dziembowski et al., 2004). Several studies also reported that unspliced mRNA is usually trapped by several components of nuclear pore complex (NPC) in the nucleus to prevent production of toxic poly-peptides (Galy et al., 2004; Palancade et al., 2005; Lewis et al., 2007). This proposed ‘perinuclear mRNP quality control’ has been recently revealed, in which endoribonuclease Swt1p is probably recruited transiently to NPCs to mediate the initiation of degradations for trapped pre-mRNP/mRNP at the perinuclear region (Skružný et al., 2009). In poly(A) binding protein (Nab2/Pab2) mutants, the observation of nuclear pre-mRNA accumulation in S. cerevisiae and Schizosaccharomyces pombe suggested that Nab2p/Pab2p binds to poly(A) tail and then recruits Rrp6p to induce specific degradation of pre-mRNA in the nucleus (Lemieux et al., 2011; Schmid et al., 2012).
Turnover of mRNAs in the nucleus is relatively slow and is mainly contributed by 5’-3’ degradation, but nucleus-retained mRNAs or pre-mRNAs are degraded quicker through the 3’-5’ degradation pathway, though Rat1p (5’-3’ degradation) is involved as well (Bousquet-Antonelli et al., 2000; Das et al., 2003; Kufel et al., 2004). More importantly, Bousquet-Antonelli et al. (2000) discovered that some spliced mRNAs are more abundant in exosome mutants than in wild-type cells. This clearly indicates that some pre-mRNAs destined to be degraded by exosome could be converted into mature mRNAs. It means that transcripts, at least a small proportion of them, have the competition between the entry of degradation and being correctly spliced to be mRNAs in the transcript pool. Furthermore, it hints that nuclear mRNA degradation induced by RNA retention might associate with a chromosome, or at least with RNP II transcription. Rougemaille et al. (2008) focusing on the THO complex, which is a multi-protein complex recruited to transcriptional machinery, come to the same conclusion that mRNP intermediate formation contains nuclear pore components and polyadenylation factors in association with chromatin. It links the quality control pathway at the perinuclear mRNP with the chromatin/transcription site, though how the THO complex functions in this link is still mysterious.
Recently, Volanakis et al. (2013) suggested that spliceosome mediates mRNA degradation in the nucleus. Transcriptome analysis of SmD1 (presented in the Sm complex, a component of spliceosome) associated RNAs identified many non-intronic transcripts. More interestingly, some of them were managed to be spliced to produce unstable mRNAs which are degraded by nuclear exosome and Rat1p. Although it is still not known whether it happened at the transcription site or another compartment in the nucleus, this spliceosome-mediated mRNA decay (SMD) plays an important and new role in yeast to minimize over-expression of proteins which are deleterious to cells.
3.3 mRNA retention at transcription site caused by 3’ end processing/RNA release
The competition is also kinetically presented in the balance of 3’ end processing and nuclear mRNA degradation. As an example, depletion of Rrp6p partially rescues the growth defect of PAP1 mutant (PAP1 encoding the RNA poly(A) polymerase), suggesting that defective polyadenylation is one of the results of mRNA degradation in the nucleus (Burkard and Butler, 2000). The same results are found in several mutations of 3’ end processing factors, Rna14p and Rna15p beside Pap1p (Minvielle-Sebastia et al., 1991; Libri et al., 2002; Torchet et al., 2002; Milligan et al., 2005). All of these studies lead us to conclude the presence of a balance between polyadenylation/RNA release and mRNA degradation. It means that transcripts are protected by poly(A) tail, which is processed by Pap1p in a normal situation, but quickly removed by exosome in the situation of aberrant 3’ end processing. This phenomenon is what exactly happened in some gene regulations. One example is the auto-regulation of NAB2 mRNA: at 3’ UTR of NAB2 mRNA, 26 consecutive adenosines (A26) compromised NAB2 mRNA 3’ end processing and induced degradation by Exo11 when excessive Nab2p bound to A26 in yeast cells (Roth et al., 2005). An additional example is that HTB1 mRNA might have a downstream unit in its transcription termination region, which could facilitate 3’ end processing factors and exosome to drastically regulate HTB1 mRNA levels when yeast cell cycles changed from S phase to G2 phase (Canavan and Bond, 2007). More interestingly, it seems that the poly(A) tail itself is required for RNA release from the transcription site by using a self-cleaving hammerhead ribozyme to eliminate normal polyadenylation. The efficient RNA release of the non-poly(A) tail mRNA occurred only if an artificial poly(A) tail was introduced at the upstream of the ribozyme cleavage site (Dower et al., 2004). Also, deletion of PAB1, which encodes poly(A) binding protein, shuttled between cytoplasm and nucleus, and PAN, which encodes poly(A) ribonuclease to trim the poly(A) tail to a proper length for mRNA export, caused the exosome-dependent nucleus retention (Dunn et al., 2005). This suggests that proper poly(A) signal/length might be the final processing step to allow mRNA release from the transcription site to be exported (Davis and Shi, 2014).
4 Bi-directional transcription and RNA decay
Transcriptome analysis unveiled that the pervasive transcription occurs in animal and yeast cells with defective exosome alleles (Jacquier, 2009). In animal cells many different RNAs have been discovered by using different techniques. However, all of them have similar distribution in the genome, either an expression peak around 50 nt downstream of transcription start site (TSS) in the same orientation as the gene expression or 250 nt upstream of TSS in a divergent transcription (Kapranov et al., 2007; Core et al., 2008; Seila et al., 2008; Jacquier, 2009). They are short-lived RNA molecules, and their 5’ ends are capped with a different mechanism, unlike normal mRNA (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009). One of the most important conclusions from these transcriptome studies in yeast and animal cells is that many promoters seem to be divergently transcribed, termed as ‘bi-directional transcription’. One extensively studied example relates to cryptic unstable transcripts (CUTs), which has been proved to be a kind of promoter-associated non-coding RNA (ncRNA) in yeast (Davis and Ares, 2006). This new type of RNA is identified only in inactivation of the function of the exosome or TRAMP complex, but not measured in wild-type cells (Arigo et al., 2006; Davis and Ares, 2006). CUTs usually are 200–600 nt long ncRNA with 5’-cap. Recently, another type of ncRNA, stable unannotated transcripts (SUTs), has been identified (Xu et al., 2009). Actually, SUTs do not have much difference from CUTs, except that SUTs are more stable and their lengths are on average longer. How CUTs/SUTs are exactly produced is still mysterious, but presumably during transcription initiation the specificity of transcription polarity is not well recognized, so that RNP II produces transcripts in two directions. The ‘polarity of transcription’ might be supported by co/post-transcriptional quality control to remove the cryptic targets. In animals, it has been suggested that bi-directional transcription occurs in transcription initiation where the transcribed regions of chromatin are modified with acetylation of Histone 3 (Preker et al., 2008).
The major difference between CUTs and encoding transcripts is 3’ end processing. In other words, probably the aberrant 3’ end RNP/processing complex induces the degradation of CUTs. It is well known that CUTs usually contain sequences, which locate less than 900 nt from TSS, recognized by Nrd1p and Nab3p, the components of the NNS complex (Gudipati et al., 2008). The recruitment of Sen1p possibly triggers the transcription termination by dissociating the elongation complex. After termination, CUTs are polyadenylated by Trf4p (poly(A) polymerase, a component of TRAMP complex), and degraded quickly by exosome (Fig. 2) (Porrua and Libri, 2013).
5 Conclusions and perspectives
Nuclear mRNA degradations in eukaryotes are complicated, multifaceted, but elaborated processes. They likely couple with every step of mRNA processing, including transcription initiation, RNP II pausing, capping, splicing, 3’ end processing, and mRNA export. Currently, we know that nuclear mRNA degradation is ubiquitous since transcription is pervasive. However, the important physiological function of mRNA degradation/quality control pathways is still to be addressed. The intrinsic mechanisms are yet to be understood from research. Furthermore, how was the defective RNA recognized by the exosome? Is the exosome associated with every RNA molecule but only degrades defective RNAs, or are the defective RNAs recognized by different mechanisms but degraded by nuclear exosome and Rat1p?
Compliance with ethics guidelines
Han LIU, Min LUO, and Ji-kai WEN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009. Post-transcriptional processing generates a diversity of 5’-modified long and short RNAs. Nature, 457(7232):1028–1032. [doi:10.1038/nature07759]
Amberg, D.C., Goldstein, A.L., Cole, C.N., 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev., 6(7):1173–1189. [doi:10.1101/gad.6.7.1173]
Arigo, J.T., Eyler, D.E., Carroll, K.L., et al., 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell, 23(6):841–851. [doi:10.1016/j.molcel.2006.07.024]
Babitzke, P., Kushner, S.R., 1991. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. PNAS, 88(1):1–5. [doi:10.1073/pnas.88.1.1]
Bousquet-Antonelli, C., Presutti, C., Tollervey, D., 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102(6):765–775. [doi:10.1016/S0092-8674(00)00065-9]
Brannan, K., Kim, H., Erickson, B., et al., 2012. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell, 46(3):311–324. [doi:10.1016/j. molcel.2012.03.006]
Brogna, S., Wen, J., 2009. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol., 16(2):107–113. [doi:10.1038/nsmb.1550]
Burkard, K.T., Butler, J.S., 2000. A nuclear 3’-5’ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol., 20(2):604–616. [doi:10.1128/MCB.20.2.604-616. 2000]
Canavan, R., Bond, U., 2007. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucl. Acids Res., 35(18):6268–6279. [doi:10.1093/nar/gkm691]
Chekulaeva, M., Filipowicz, W., 2009. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol., 21(3):452–460. [doi:10. 1016/j.ceb.2009.04.009]
Chen, C.Y., Gherzi, R., Ong, S.E., et al., 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell, 107(4):451–464. [doi:10.1016/S0092-8674(01)00578-5]
Core, L.J., Waterfall, J.J., Lis, J.T., 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science, 322(5909):1845–1848. [doi:10.1126/science.1162228]
Das, B., Guo, Z., Russo, P., et al., 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol., 20(8):2827–2838. [doi:10. 1128/MCB.20.8.2827-2838.2000]
Das, B., Butler, J.S., Sherman, F., 2003. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol., 23(16):5502–5515. [doi:10.1128/MCB.23.16. 5502-5515.2003]
Davis, C.A., Ares, M.Jr., 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. PNAS, 103(9):3262–3267. [doi:10.1073/pnas.0507783103]
Davis, R., Shi, Y., 2014. The polyadenylation code: a unified model for the regulation of mRNA alternative polyadenylation. J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol.), 15(5):429–437. [doi:10.1631/jzus.B1400076]
Doma, M.K., Parker, R., 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature, 440(7083):561–564. [doi:10.1038/nature04530]
Dower, K., Kuperwasser, N., Merrikh, H., et al., 2004. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA, 10(12):1888–1899. [doi:10.1261/rna.7166704]
Dunn, E.F., Hammell, C.M., Hodge, C.A., et al., 2005. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev., 19(1):90–103. [doi:10. 1101/gad.1267005]
Dziembowski, A., Ventura, A.P., Rutz, B., et al., 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J., 23(24):4847–4856. [doi:10.1038/sj.emboj.7600482]
Dziembowski, A., Lorentzen, E., Conti, E., et al., 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol., 14(1):15–22. [doi:10.1038/nsmb1184]
Even, S., Pellegrini, O., Zig, L., et al., 2005. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucl. Acids Res., 33(7):2141–2152. [doi:10.1093/nar/gki505]
Galy, V., Gadal, O., Fromont-Racine, M., et al., 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell, 116(1):63–73. [doi:10.1016/ S0092-8674(03)01026-2]
Geerlings, T.H., Vos, J.C., Raue, H.A., 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5’→3’ exonucleases. RNA, 6(12):1698–1703. [doi:10.1017/S1355838200001540]
Gonatopoulos-Pournatzis, T., Cowling, V.H., 2014. Cap-binding complex (CBC). Biochem. J., 458(1):185.
Grigull, J., Mnaimneh, S., Pootoolal, J., et al., 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol., 24(12):5534–5547. [doi:10.1128/MCB.24.12.5534-5547. 2004]
Gudipati, R.K., Villa, T., Boulay, J., et al., 2008. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol., 15(8):786–794. [doi:10.1038/nsmb.1460]
Gudipati, R.K., Xu, Z.Y., Lebreton, A., et al., 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell, 48(3):409–421. [doi:10.1016/ j.molcel.2012.08.018]
Halbach, F., Reichelt, P., Rode, M., et al., 2013. The yeast Ski complex: crystal structure and RNA channeling to the exosome complex. Cell, 154(4):814–826. [doi:10.1016/ j.cell.2013.07.017]
Henry, Y., Wood, H., Morrissey, J.P., et al., 1994. The 5’ end of yeast 5. 8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13(10):2452–2463.
Hilleren, P., McCarthy, T., Rosbash, M., et al., 2001. Quality control of mRNA 3’-end processing is linked to the nuclear exosome. Nature, 413(6855):538–542. [doi:10. 1038/35097110]
Houseley, J., LaCava, J., Tollervey, D., 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol., 7(7):529–539. [doi:10.1038/nrm1964]
Jackson, D.A., Pombo, A., Iborra, F., 2000. The balance sheet for transcription: an analysis of nuclear RNA metabolism in mammalian cells. FASEB J., 14(2):242–254.
Jacquier, A., 2009. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet., 10(12):833–844. [doi:10.1038/ nrg2683]
Jiao, X.F., Xiang, S., Oh, C., et al., 2010. Identification of a quality-control mechanism for mRNA 5’-end capping. Nature, 467(7315):608–611. [doi:10.1038/nature09338]
Kapranov, P., Cheng, J., Dike, S., et al., 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 316(5830):1484–1488. [doi: 10.1126/science.1138341]
Kim, M., Krogan, N.J., Vasiljeva, L., et al., 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature, 432(7016):517–522. [doi:10. 1038/nature03041]
Kim, M., Vasiljeva, L., Rando, O.J., et al., 2006. Distinct pathways for snoRNA and mRNA termination. Mol. Cell, 24(5):723–734. [doi:10.1016/j.molcel.2006.11.011]
Kim, Y.K., Furic, L., Desgroseillers, L., et al., 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell, 120(2):195–208. [doi:10.1016/j.cell.2004.11.050]
Kuai, L., Das, B., Sherman, F., 2005. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. PNAS, 102(39):13962–13967. [doi:10.1073/pnas.0506518102]
Kufel, J., Bousquet-Antonelli, C., Beggs, J.D., et al., 2004. Nuclear pre-mRNA decapping and 5’ degradation in yeast require the Lsm2-8p complex. Mol. Cell. Biol., 24(21):9646–9657. [doi:10.1128/MCB.24.21.9646-9657.2004]
LaCava, J., Houseley, J., Saveanu, C., et al., 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell, 121(5):713–724. [doi:10. 1016/j.cell.2005.04.029]
Legrain, P., Rosbash, M., 1989. Some cis- and transacting mutants for splicing target pre-mRNA to the cytoplasm. Cell, 57(4):573–583. [doi:10.1016/0092-8674(89)90127-X]
Lemieux, C., Marguerat, S., Lafontaine, J., et al., 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)- binding protein. Mol. Cell, 44(1):108–119. [doi:10.1016/ j.molcel.2011.06.035]
Lewis, A., Felberbaum, R., Hochstrasser, M., 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol., 178(5):813–827. [doi:10.1083/jcb.200702154]
Li, C.H., Irmer, H., Gudjonsdottir-Planck, D., et al., 2006. Roles of a Trypanosoma brucei 5’→3’ exoribonuclease homolog in mRNA degradation. RNA, 12(12):2171–2186. [doi:10.1261/rna.291506]
Libri, D., Dower, K., Boulay, J., et al., 2002. Interactions between mRNA export commitment, 3’-end quality control, and nuclear degradation. Mol. Cell. Biol., 22(23):8254–8266. [doi:10.1128/MCB.22.23.8254-8266.2002]
Liu, Q.S., Greimann, J.C., Lima, C.D., 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell, 127(6):1223–1237. [doi:10.1016/j.cell.2006.10.037]
Lubas, M., Christensen, M.S., Kristiansen, M.S., et al., 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell, 43(4):624–637. [doi: 10.1016/j.molcel.2011.06.028]
Luke, B., Panza, A., Redon, S., et al., 2008. The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell, 32(4):465–477. [doi:10.1016/j. molcel.2008.10.019]
Malecki, M., Viegas, S.C., Carneiro, T., et al., 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J., 32(13):1842–1854. [doi: 10.1038/emboj.2013.63]
Mathy, N., Benard, L., Pellegrini, O., et al., 2007. 5’-to-3’ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5’ stability of mRNA. Cell, 129(4):681–692. [doi:10.1016/j.cell.2007.02.051]
Milligan, L., Torchet, C., Allmang, C., et al., 2005. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol., 25(22):9996–10004. [doi:10. 1128/MCB.25.22.9996-10004.2005]
Minvielle-Sebastia, L., Winsor, B., Bonneaud, N., et al., 1991. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol., 11(6):3075–3087.
Mitchell, P., Petfalski, E., Shevchenko, A., et al., 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3’→5’ exoribonucleases. Cell, 91(4):457–466. [doi:10.1016/S0092-8674(00)80432-8]
Mitchell, P., Petfalski, E., Houalla, R., et al., 2003. Rrp47p is an exosome-associated protein required for the 3’ processing of stable RNAs. Mol. Cell. Biol., 23(19):6982–6992. [doi:10.1128/MCB.23.19.6982-6992.2003]
Montero Llopis, P., Jackson, A.F., Sliusarenko, O., et al., 2010. Spatial organization of the flow of genetic information in bacteria. Nature, 466(7302):77–81. [doi:10.1038/nature 09152]
Palancade, B., Zuccolo, M., Loeillet, S., et al., 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell, 16(11):5258–5268. [doi:10.1091/mbc.E05-06-0527]
Peng, S.S., Chen, C.Y., Xu, N., et al., 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J., 17(12):3461–3470. [doi:10.1093/ emboj/17.12.3461]
Porrua, O., Libri, D., 2013. RNA quality control in the nucleus: the Angels’ share of RNA. Biochim. Biophys. Acta, 1829(6-7):604–611. [doi:10.1016/j.bbagrm.2013.02.012]
Preker, P., Nielsen, J., Kammler, S., et al., 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science, 322(5909):1851–1854. [doi: 10.1126/science.1164096]
Proudfoot, N.J., 2011. Ending the message: poly(A) signals then and now. Genes Dev., 25(17):1770–1782. [doi:10. 1101/gad.17268411]
Richard, P., Manley, J.L., 2009. Transcription termination by nuclear RNA polymerases. Genes Dev., 23(11):1247–1269. [doi:10.1101/gad.1792809]
Roth, K.M., Wolf, M.K., Rossi, M., et al., 2005. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol. Cell. Biol., 25(5):1577–1585. [doi:10. 1128/MCB.25.5.1577-1585.2005]
Rougemaille, M., Dieppois, G., Kisseleva-Romanova, E., et al., 2008. THO/Sub2p functions to coordinate 3’-end processing with gene-nuclear pore association. Cell, 135(2):308–321. [doi:10.1016/j.cell.2008.08.005]
Rutz, B., Seraphin, B., 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J., 19(8):1873–1886. [doi:10.1093/emboj/19.8.1873]
Schmid, M., Poulsen, M.B., Olszewski, P., et al., 2012. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol. Cell, 47(2):267–280. [doi:10.1016/j.molcel.2012.05.005]
Schneider, C., Kudla, G., Wlotzka, W., et al., 2012. Transcriptome-wide analysis of exosome targets. Mol. Cell, 48(3):422–433. [doi:10.1016/j.molcel.2012.08.013]
Schroeder, S.C., Zorio, D.A., Schwer, B., et al., 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell, 13(3):377–387. [doi:10.1016/S1097-2765(04)00007-3]
Schwer, B., Mao, X., Shuman, S., 1998. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucl. Acids Res., 26(9):2050–2057. [doi:10. 1093/nar/26.9.2050]
Seila, A.C., Calabrese, J.M., Levine, S.S., et al., 2008. Divergent transcription from active promoters. Science, 322(5909):1849–1851. [doi:10.1126/science.1162253]
Shahbabian, K., Jamalli, A., Zig, L., et al., 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J., 28(22):3523–3533. [doi:10. 1038/emboj.2009.283]
Shobuike, T., Sugano, S., Yamashita, T., et al., 1995. Characterization of cDNA encoding mouse homolog of fission yeast dhp1+ gene: structural and functional conservation. Nucl. Acids Res., 23(3):357–361. [doi:10.1093/nar/23.3. 357]
Shobuike, T., Tatebayashi, K., Tani, T., et al., 2001. The dhp1+ gene, encoding a putative nuclear 5’→3’ exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucl. Acids Res., 29(6):1326–1333. [doi:10.1093/ nar/29.6.1326]
Skružný, M., Schneider, C., Rácz, A., et al., 2009. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol., 7(1):e8. [doi:10.1371/journal.pbio. 1000008]
Stevens, A., Poole, T.L., 1995. 5’-Exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5’-exonuclease-1. J. Biol. Chem., 270(27):16063–16069. [doi:10.1074/jbc.270. 27.16063]
Taraseviciene, L., Miczak, A., Apirion, D., 1991. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol. Microbiol., 5(4):851–855. [doi:10.1111/j.1365-2958.1991.tb00758.x]
Tomecki, R., Kristiansen, M.S., Lykke-Andersen, S., et al., 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J., 29(14):2342–2357. [doi:10.1038/emboj.2010. 121]
Torchet, C., Bousquet-Antonelli, C., Milligan, L., et al., 2002. Processing of 3’-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell, 9(6):1285–1296. [doi:10.1016/S1097-2765(02)00544-0]
Vanacova, S., Stefl, R., 2007. The exosome and RNA quality control in the nucleus. EMBO Rep., 8(7):651–657. [doi:10.1038/sj.embor.7401005]
van Hoof, A., Frischmeyer, P.A., Dietz, H.C., et al., 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science, 295(5563):2262–2264. [doi:10.1126/science.1067272]
Volanakis, A., Passoni, M., Hector, R.D., et al., 2013. Spliceosome-mediated decay (SMD) regulates expression of nonintronic genes in budding yeast. Genes Dev., 27(18):2025–2038. [doi:10.1101/gad.221960.113]
Wagschal, A., Rousset, E., Basavarajaiah, P., et al., 2012. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell, 150(6):1147–1157. [doi:10.1016/j.cell.2012.08.004]
Wang, Y., Liu, C.L., Storey, J.D., et al., 2002. Precision and functional specificity in mRNA decay. PNAS, 99(9):5860–5865. [doi:10.1073/pnas.092538799]
West, S., Gromak, N., Proudfoot, N.J., 2004. Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature, 432(7016):522–525. [doi:10.1038/nature03035]
Xiang, S., Cooper-Morgan, A., Jiao, X.F., et al., 2009. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature, 458(7239):784–788. [doi:10.1038/nature07731]
Xu, Z., Wei, W., Gagneur, J., et al., 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature, 457(7232):1033–1037. [doi:10.1038/nature07728]
Xue, Y., Bai, X., Lee, I., et al., 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol., 20(11):4006–4015. [doi:10.1128/ MCB.20.11.4006-4015.2000]
Yang, E., van Nimwegen, E., Zavolan, M., et al., 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res., 13(8):1863–1872.
Yoo, E.J., Jin, Y.H., Jang, Y.K., et al., 2000. Fission yeast Hrp1, a chromodomain ATPase, is required for proper chromosome segregation and its overexpression interferes with chromatin condensation. Nucl. Acids Res., 28(9):2004–2011. [doi:10.1093/nar/28.9.2004]
Zhai, L.T., Xiang, S., 2014. mRNA quality control at the 5’ end. J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol.), 15(5):438–443. [doi:10.1631/jzus.B1400070]
Author information
Authors and Affiliations
Corresponding author
Additional information
Project supported by the Talented Scientist Program from South China Agricultural University (No. 4600-K14013) and the National Natural Science Foundation of China (No. 81301901)
The two authors contributed equally to this work
Rights and permissions
About this article
Cite this article
Liu, H., Luo, M. & Wen, Jk. mRNA stability in the nucleus. J. Zhejiang Univ. Sci. B 15, 444–454 (2014). https://doi.org/10.1631/jzus.B1400088
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1400088