Abstract
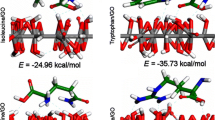
In an attempt to understand the structure and dynamics of ssDNA on graphene based surfaces, we performed all-atom implicit solvent molecular dynamics simulations of ssDNA on graphene and graphene oxide (GO) surfaces. Simulations indicate that adsorption of poly(A), poly(T) and poly (AT) have similar mechanisms of adsorption to free standing graphitic flakes, which are governed by a surface oxygen content. Specifically, higher oxygen content of a surface leads to decrease in persistence length of ssDNA. However, the role of DNA sequence on the physisorption mechanism is minimal.
Similar content being viewed by others
References
M.E. Hogan, R.H. Austin, “Importance of DNA Stiffness in Protein DNA-Binding Specificity” Nature 1987, 329, 263–266.
J.J. Gooding, G.C. King, “Nucleic acid biosensors based upon surface-assembled monolayers: exploiting and enhancing materials properties” Journal of Materials Chemistry 2005, 15, 4876–4880.
M.M. Lozano, CD. Starkel, M.L. Longo, “Vesicles Tethered to Microbubbles by Hybridized DNA Oligonucleotides: Flow Cytometry Analysis of This New Drug Delivery Vehicle Design” Langmuir 2010, 26, 8517–8524.
N.K. Li, H.S. Kim, J.A. Nash, M. Lim, Y.G. Yingling, “Progress in molecular modelling of DNA materials” Molecular Simulation 2014, 40, 777–783.
A. Singh, H. Eksiri, Y.G. Yingling, “Theoretical Perspective on Properties of DNA-Functionalized Surfaces” Journal of Polymer Science Part B-Polymer Physics 2011, 49, 1563–1568.
Y. He, B.N. Jiao, H.W. Tang, “Interaction of single-stranded DNA with graphene oxide: fluorescence study and its application for S1 nuclease detection” Rsc Advances 2014, 4, 18294–18300.
A. Singh, S. Snyder, L. Lee, A.P.R. Johnston, F. Caruso, Y.G. Yingling, “Effect of Oligonucleotide Length on the Assembly of DNA Materials: Molecular Dynamics Simulations of Layer-by-Layer DNA Films” Langmuir 2010, 26, 17339–17347.
Y.W. Zhu, S. Murali, W.W. Cai, X.S. Li, J.W. Suk, J.R. Potts, R.S. Ruoff, “Graphene and Graphene Oxide: Synthesis, Properties, and Applications” Advanced Materials 2010, 22, 3906–3924.
J.L. Chen, X.G. Wang, C.Q. Dai, S.D. Chen, Y.S. Tu, “Adsorption of GA module onto graphene and graphene oxide: A molecular dynamics simulation study” Physica E-Low-Dimensional Systems & Nanostructures 2014, 62, 59–63.
L. Baweja, K. Balamurugan, V. Subramanian, A. Dhawan, “Hydration Patterns of Graphene-Based Nanomaterials (GBNMs) Play a Major Role in the Stability of a Helical Protein: A Molecular Dynamics Simulation Study” Langmuir 2013, 29, 14230–14238.
X.T. Sun, Z.W. Feng, T.J. Hou, Y.Y. Li, “Mechanism of Graphene Oxide as an Enzyme Inhibitor from Molecular Dynamics Simulations” Acs Applied Materials & Interfaces 2014,6,7153–7163.
T.J. Macke, D.A. Case, “Modeling unusual nucleic acid structures” Molecular Modeling of Nucleic Acids 1998, 682, 379–393.
D. Case, T. Darden, T. Cheatham III, C. Simmerling, J. Wang, R. Duke, R. Luo, R. Walker, W. Zhang, K. Merz, “AMBER 12” University of California, San Francisco 2012.
K. Range, E. Mayaan, L.J. Maher, D.M. York, “The contribution of phosphate-33hosphate repulsions to the free energy of DNA bending” Nucleic Acids Research 2005, 33, 1257–1268.
J.M. Wang, P. Cieplak, P.A. Kollman, “How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules?” Journal of Computational Chemistry 2000, 21, 1049–1074.
W. Humphrey, A. Dalke, K. Schulten, “VMD: Visual molecular dynamics” Journal of Molecular Graphics & Modelling 1996, 14, 33–38.
J.M. Wang, R.M. Wolf, J.W. Caldwell, P.A. Kollman, D.A. Case, “Development and testing of a general amber force field” Journal of Computational Chemistry 2004, 25, 1157–1174.
D. Stauffer, N. Dragneva, W.B. Floriano, R.C. Mawhinney, G. Fanchini, S. French, O. Rubel, “An atomic charge model for graphene oxide for exploring its bioadhesive properties in explicit water” Journal of Chemical Physics 2014,141.
H.J.C. Berendsen, J.P.M. Postma, W.F. Vangunsteren, A. Dinola, J.R. Haak, “Molecular-Dynamics with Coupling to an External Bath” Journal of Chemical Physics 1984, 81, 3684–3690.
M. Rubinstein, R. Colby, Polymers Physics. Oxford: 2003.
D. Bashford, D.A. Case, “Generalized born models of macromolecular solvation effects” Annual Review of Physical Chemistry 2000, 51, 129–152.
A.K. Manna, S.K. Pati, “Theoretical understanding of single-stranded DNA assisted dispersion of graphene” Journal of Materials Chemistry B 2013, 1, 91–100.
H.S. Kim, S.H. Ha, L. Sethaphong, Y.M. Koo, Y.G. Yingling, “The relationship between enhanced enzyme activity and structural dynamics in ionic liquids: a combined computational and experimental study” Physical Chemistry Chemical Physics 2014,16, 2944–2953.
H.S. Kim, R. Pani, S.H. Ha, Y.M. Koo, Y.G. Yingling, “The role of hydrogen bonding in water-mediated glucose solubility in ionic liquids” Journal of Molecular Liquids 2012, 166, 25–30.
D.R. Roe, T.E. Cheatham, “PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data” Journal of Chemical Theory and Computation 2013, 9, 3084–3095.
G.B. McGaughey, M. Gagne, A.K. Rappe, “pi-stacking interactions - Alive and well in proteins” Journal of Biological Chemistry 1998, 273, 15458–15463.
C.A. Hunter, J. Singh, J.M. Thornton, “Pi-Pi-Interactions - the Geometry and Energetics of Phenylalanine Phenylalanine Interactions in Proteins” Journal of Molecular Biology 1991, 218, 837–846.
A.N. Round, M. Berry, T.J. McMaster, S. Stall, D. Gowers, A.P. Corfield, M.J. Miles, “Hetero3eneity and persistence length in human ocular mucins” Biophysical Journal 2002, 83,1661–1670.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kim, H.S., Huang, S.M. & Yingling, Y.G. Sequence dependent interaction of single stranded DNA with graphitic flakes: atomistic molecular dynamics simulations. MRS Advances 1, 1883–1889 (2016). https://doi.org/10.1557/adv.2016.91
Published:
Issue Date:
DOI: https://doi.org/10.1557/adv.2016.91