Abstract
Background
A number of long non-coding RNAs (lncRNAs) have been found to be involved in tumor progression and associated with disease prognoses in various types of cancer. Our study identified a novel three-lncRNA signature to predict survival of head and neck squamous cell caner (HNSCC) patients.
Methods
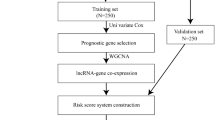
We utilized The Cancer Genome Atlas (TCGA) cohort to screen out overall survival (OS)-associated lncRNAs in HNSCC and further developed a model to identify a lncRNA signature for evaluating disease status and prognosis. The lncRNA signature was then validated in HNSCC patients from our Fudan University Shanghai Cancer Center (FUSCC) cohort.
Results
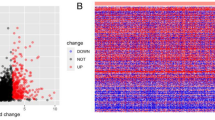
LINC02434, AL139327.2, and AC126175.1 were identified by multivariable Cox regression analyses of independent risk factors for deceased status. We built a risk score model based on the three-lncRNA signature using coefficient of multivariable Cox regression and expression value of the three lncRNAs. The high-risk signature score was significantly associated with decreased OS in both the TCGA cohort and the FUSCC cohort. The high-risk group had worse overall survival than the low-risk group in TCGA cohort. To further validate the robustness of three-lncRNA signature risk score model developed in the TCGA dataset, the performance of risk score also evaluated in our institute FUSCC cohort. Additionally, the signature score showed a positive correlation with aggressive outcomes of HNSCC, such as III/IV stage, TP53 mutation, and PI3KCA mutation. The gene set enrichment analysis indicates that the risk score is associated with cancer metastasis-related pathways. Several cancer-related pathways, such as epithelial mesenchymal transition, TNFα signaling via NF-κB, MYC targets, and angiogenesis.
Conclusions
The three-lncRNA signature could provide a novel prediction insight into the prognosis of HNSCC patients. The three-lncRNA signature was identified as a predictor of poor prognoses in HNSCC, which may serve as a potential therapeutic target.






Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Katiyar S. Emerging phytochemicals for the prevention and treatment of head and neck cancer. Molecules. 2016;21:1610.
Fleming J, Woo J, Moutasim K, Mellone M, Frampton S, Mead A, et al. HPV, tumour metabolism and novel target identification in head and neck squamous cell carcinoma. Br J Cancer. 2019;120:356–67.
Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18:269.
Foy J, Bazire L, Ortiz-Cuaran S, Deneuve S, Kielbassa J, Thomas E, et al. A 13-gene expression-based radioresistance score highlights the heterogeneity in the response to radiation therapy across HPV-negative HNSCC molecular subtypes. BMC Med. 2017;15:1–13.
Lee DJ, Eun Y, Rho YS, Kim EH, Yim SY, Kang SH, et al. Three distinct genomic subtypes of head and neck squamous cell carcinoma associated with clinical outcomes. Oral Oncol. 2018;85:44–51.
Leemans C, Braakhuis B, Brakenhoff R. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22.
Ahmad MK, Mitchell G, Maite H, Manuel G, Arjun R, Dianali RM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72.
Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:1–13.
Wu Q, Meng W, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018; 233:6750–7.
Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–109.
Ma B, Liao T, Wen D, Dong C, Zhou L, Yang S, et al. Long intergenic non-coding RNA 271 is predictive of a poorer prognosis of papillary thyroid cancer. Sci Rep-UK. 2016;6:1–13.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Schmitt AM, Chang HY. Long noncoding RNAs: at the intersection of cancer and chromatin biology. CSH Perspect Med. 2017;7:a026492.
Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi Q, et al. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer. 2018;17:1–6.
Zhang L, Kang W, Lu X, Ma S, Dong L, Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle (Georgetown, Tex.). 2018;17:1886–900.
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–24.
Chiu-Lien H, Ling-Yu W, Yen-Ling Y, Hong-Wu C, Shiv S, Gyorgy P, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. 2014;111:18697–702.
Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343–54.
Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: a new regulator in cancer signaling. Cancer Lett. 2018;419:27–9.
Liao M, Liao W, Xu N, Li B, Liu F, Zhang S, et al. LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2. Ebiomedicine. 2019;41:200–13.
Nohata N, Abba M, Gutkind JS. Unraveling the oral cancer lncRNAome: Identification of novel lncRNAs associated with malignant progression and HPV infection. Oral Oncol. 2016;59:58–66.
Cao W, Liu J, Liu Z, Wang X, Han Z, Ji T, et al. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101.
Liu G, Zheng J, Zhuang L, Lv Y, Zhu G, Pi L, et al. A prognostic 5-lncRNA expression signature for head and neck squamous cell carcinoma. Sci Rep-UK. 2018;8:1–13.
Zhang Z, Zhao L, Chai L, Zhou S, Wang F, Wei Y, et al. Seven LncRNA-mRNA based risk score predicts the survival of head and neck squamous cell carcinoma. Sci Rep-UK. 2017;7:1–9.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–50.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England). 2010;26:139–40.
Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10:1–16.
Du E, Mazul AL, Farquhar D, Brennan P, Anantharaman D, Abedi-Ardekani B, et al. Long-term survival in head and neck cancer: impact of site, stage, smoking, and human papillomavirus status. Laryngoscope. 2019;129:2506–13.
Solomon B, Young RJ, Rischin D. Head and neck squamous cell carcinoma: genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol. 2018;52:228–40.
Read ML, Modasia B, Fletcher A, Thompson RJ, Brookes K, Rae PC, et al. PTTG and PBF Functionally Interact with p53 and predict overall survival in head and neck cancer. Cancer Res. 2018;78:5863–76.
Jou A, Hess J. Epidemiology and molecular biology of head and neck cancer. Oncol Res Treat. 2017;40:328–32.
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–903.
Chen Q, Hu C, Sun K, Lang Y. LncRNA NR027113 promotes malignant progression of gastric carcinoma via EMT signaling pathway. Eur Rev Med Pharmacol. 2019;23:4746–55.
Zhang Z, Lin G, Yan Y, Li X, Hu Y, Wang J, et al. Transmembrane TNF-alpha promotes chemoresistance in breast cancer cells. Oncogene. 2018;37:3456–70.
Stine Z, Walton Z, Altman B, Hsieh A, Dang C. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–39.
Acknowledgement
The authors thank Yifan Yang for her help in this study and TCGA platform for cancer genomics data sets. The research was supported by Shanghai Anticancer Association (SACA-AX106 to Yu Wang) and Shanghai Anticancer Association (SACA-AX106 to Yu Wang and SACA-CY19B01 to Ben Ma).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosures
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
. The enrichment biological cancer related pathway of each lncRNA based on the GSEA of TCGA cohort. (A) The enrichment biological cancer related pathway of AC126175.1 based on the GSEA of TCGA cohort. (B) The enrichment biological cancer related pathway of AL139327.21 based on the GSEA of TCGA cohort. (C). The enrichment biological cancer related pathway of LINC02434 based on the GSEA of TCGA cohort.
Supplementary Figure 2
. Kaplan-Meier curves for early stage (stage I/II) and advanced stage (stage III/IV). (A) Kaplan-Meier curves for early stage (stage I/II). (B) Kaplan-Meier curves for advanced stage (stage III/IV).
Rights and permissions
About this article
Cite this article
Jiang, H., Ma, B., Xu, W. et al. A Novel Three-lncRNA Signature Predicts the Overall Survival of HNSCC Patients. Ann Surg Oncol 28, 3396–3406 (2021). https://doi.org/10.1245/s10434-020-09210-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-020-09210-1