Abstract
Background
Circulating tumor DNA (ctDNA) is a promising technology for treatment selection, prognostication, and surveillance after definitive therapy. Its use in the perioperative setting for patients with metastatic disease has not been well studied. We characterize perioperative plasma ctDNA and its association with progression-free survival (PFS) in patients undergoing surgery for peritoneal metastases.
Patients and Methods
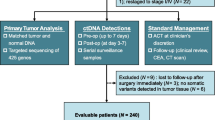
We recruited 71 patients undergoing surgery for peritoneal metastases and evaluated their plasma with a targeted 73-gene ctDNA next-generation sequencing test before and after surgery. The association between perioperative ctDNA, as well as other patient factors, and PFS was evaluated by Cox regression.
Results
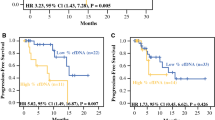
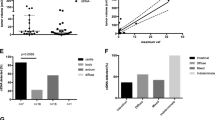
ctDNA was detectable in 28 patients (39.4%) preoperatively and in 37 patients (52.1%) postoperatively. Patients with high ctDNA [maximum somatic variant allele fraction (MSVAF) > 0.25%] had worse PFS than those with low MSVAF (< 0.25%) in both the pre- and postoperative settings (median 4.8 vs. 19.3 months, p < 0.001, and 9.2 vs.15.0 months, p = 0.049, respectively; log-rank test). On multivariate analysis, high-grade histology [hazard ratio (HR) 3.42, p = 0.001], incomplete resection (HR 2.35, p = 0.010), and high preoperative MSVAF (HR 3.04, p = 0.001) were associated with worse PFS. Patients with new postoperative alterations in the context of preoperative alteration(s) also had a significantly shorter PFS compared with other groups (HR 4.28, p < 0.001).
Conclusions
High levels of perioperative ctDNA and new postoperative ctDNA alterations in the context of preoperative alterations predict worse outcomes in patients undergoing resection for peritoneal metastases. This may highlight a role for longitudinal ctDNA surveillance in this population.


Similar content being viewed by others
References
Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7(9):1006–1017.
Ikeda S, Schwaederle M, Mohindra M, Fontes Jardim DL, Kurzrock R. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J Hematol Oncol. 2018;11(1):76.
Kato S, Okamura R, Baumgartner JM, et al. Analysis of circulating tumor DNA and clinical correlates in patients with esophageal, gastroesophageal junction, and gastric adenocarcinoma. Clin Cancer Res. 2018;24(24):6248–6256.
Kato S, Okamura R, Mareboina M, et al. Revisiting epidermal growth factor feceptor (EGFR) amplification as a target for anti-EGFR therapy: analysis of cell-free circulating tumor DNA in patients with advanced malignancies. JCO Precis Oncol. 2019. https://doi.org/10.1200/PO.18.00180.
Kato S, Schwaederle MC, Fanta PT, et al. Genomic assessment of blood-derived circulating tumor DNA in patients with colorectal cancers: correlation with tissue sequencing, therapeutic response, and survival. JCO Precis Oncol. 2019. https://doi.org/10.1200/PO.18.00158.
Riviere P, Fanta PT, Ikeda S, Baumgartner J, Heestand GM, Kurzrock R. The mutational landscape of gastrointestinal malignancies as reflected by circulating tumor DNA. Mol Cancer Ther. 2018;17(1):297–305.
Shatsky R, Parker BA, Bui NQ, et al. Next-Generation sequencing of tissue and circulating tumor DNA: the UC San Diego Moores Center for personalized cancer therapy experience with breast malignancies. Mol Cancer Ther. 2019;18(5):1001–1011.
Tjensvoll K, Lapin M, Buhl T, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. 2016;10(4):635–643.
Mardinian K, Okamura R, Kato S, Kurzrock R. Temporal and spatial effects and survival outcomes associated with concordance between tissue and blood KRAS alterations in the pan-cancer setting. Int J Cancer. 2020;146(2):566–576.
Baumgartner JM, Raymond VM, Lanman RB, et al. Preoperative circulating tumor DNA in patients with peritoneal carcinomatosis is an independent predictor of progression-free survival. Ann Surg Oncol. 2018;25(8):2400–2408.
Scholer LV, Reinert T, Orntoft MW, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–5445.
Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990.
Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):827.
Pu WY, Zhang R, Xiao L, et al. Prediction of cancer progression in a group of 73 gastric cancer patients by circulating cell-free DNA. BMC Cancer. 2016;16(1):943.
Sugarbaker PH, Stuart OA, Vidal-Jove J, Pessagno AM, DeBruijn EA. Pharmacokinetics of the peritoneal-plasma barrier after systemic mitomycin C administration. Cancer Treat Res. 1996;82:41–52.
Chua TC, Moran BJ, Sugarbaker PH, et al. Early- and long-term outcome data of patients with pseudomyxoma peritonei from appendiceal origin treated by a strategy of cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. J Clin Oncol. 2012;30(20):2449–2456.
Yan TD, Deraco M, Baratti D, et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for malignant peritoneal mesothelioma: multi-institutional experience. J Clin Oncol. 2009;27(36):6237–6242.
Franko J, Ibrahim Z, Gusani NJ, Holtzman MP, Bartlett DL, Zeh HJ, 3rd. Cytoreductive surgery and hyperthermic intraperitoneal chemoperfusion versus systemic chemotherapy alone for colorectal peritoneal carcinomatosis. Cancer. 2010;116(16):3756–3762.
Verwaal VJ, Bruin S, Boot H, van Slooten G, van Tinteren H. 8-year follow-up of randomized trial: cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy in patients with peritoneal carcinomatosis of colorectal cancer. Ann Surg Oncol. 2008;15(9):2426–2432.
Baumgartner JM, Tobin L, Heavey SF, Kelly KJ, Roeland EJ, Lowy AM. Predictors of progression in high-grade appendiceal or colorectal peritoneal carcinomatosis after cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Ann Surg Oncol. 2015;22(5):1716–1721.
Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10(10):e0140712.
Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24(15):3539–3549.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247.
Wickham H. Easily Install and Load the “Tidyverse.” tidyverse;2017.
Maron SB, Chase LM, Lomnicki S, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res. 2019;25(23):7098–7112.
Khier S, Lohan L. Kinetics of circulating cell-free DNA for biomedical applications: critical appraisal of the literature. Future Sci OA. 2018;4(4):FSO295.
Yang YC, Wang D, Jin L, et al. Circulating tumor DNA detectable in early- and late-stage colorectal cancer patients. Biosci Rep. 2018. https://doi.org/10.1042/BSR20180322.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Financial support
Funded in part by the National Cancer Institute (grant P30 CA023100) and the Joan and Irwin Jacobs Fund philanthropic fund. Study also funded in part by the Guardant Health (Guardant Health, Inc., Redwood City, CA). The project described was partially supported by the National Institutes of Health (grant TL1TR001443 of CTSA funding beginning August 13, 2015 and beyond). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosures
Razelle Kurzrock discloses stock and other equity interests (IDbyDNA, CureMatch, Inc., and Soluventis); consulting or advisory role (Gaido, LOXO, X-Biotech, Actuate Therapeutics, Roche, NeoMed, Soluventis, and Pfizer); speaker’s fee (Roche); research Fundingf [Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Guardant Health, Grifols, Konica Minolta, DeBiopharm, Boerhringer Ingelheim, and OmniSeq (all institutional)]; board member (CureMatch, Inc). Paul Riviere discloses consulting fees from Peptide Logic, LLC. Richard Lanman is an employee of Guardant Health, Inc.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baumgartner, J.M., Riviere, P., Lanman, R.B. et al. Prognostic Utility of Pre- and Postoperative Circulating Tumor DNA Liquid Biopsies in Patients with Peritoneal Metastases. Ann Surg Oncol 27, 3259–3267 (2020). https://doi.org/10.1245/s10434-020-08331-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-020-08331-x