Abstract
Background
Neo-adjuvant chemotherapy (NaCT) facilitates complete surgical resection in locally advanced breast cancer. Due to its association with improved outcome, complete pathologic response (pCR) to neo-adjuvant treatment has been accepted as a surrogate for long-term outcome in clinical trials of human epidermal growth factor receptor 2 (HER2)-positive, triple-negative, or luminal B breast cancer patients. In contrast, NaCT is effective in only ~ 7–10% of estrogen receptor (ER)-positive, HER2-negative disease. Response biomarkers would enable such patients to be selected for NaCT.
Methods
Two commercially available breast cancer prognostic signatures [12-gene molecular score (MS) and the 21-gene Recurrence Score (RS)] were compared in their ability to predict pCR to NaCT in ER-positive, HER2-negative breast cancer in six public RNA expression microarray data sets. Scores were approximated according to published algorithms and analyzed by logistic regression.
Results
Expression data were available for 764 ER-positive, HER2-negative breast cancer samples, including 59 patients with pCR. The two scores were well correlated. Either score was a significant predictor of pCR (12-gene MS p = 9.4 × 10−5; 21-gene RS p = 0.0041). However, in a model containing both scores, the 12-gene MS remained significant (p = 0.0079), while the 21-gene RS did not (p = 0.79).
Conclusions
In this microarray study, two commercial breast cancer prognostic scores were significant predictors of response to NaCT. In direct comparison, the 12-gene MS outperformed the 21-gene RS as a predictive marker for NaCT. Considering pCR as surrogate for improved survival, these results support the ability of both scores to predict chemotherapy sensitivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Over the last decade preoperative systemic therapy has become standard treatment for locally advanced breast cancer and is a treatment option for many patients with early-stage breast cancer.1 Neo-adjuvant chemotherapy (NaCT) frequently results in down-staging of both primary tumor and local nodes, rendering previously inoperable tumors amenable to surgical resection. Complete pathological response (pCR) to NaCT is a surrogate marker for improved survival of patients with human epidermal growth factor 2 (HER2)-positive, triple-negative, or luminal B breast cancer.2 In addition, neo-adjuvant treatment enables assessment of sensitivity to specific drugs in vivo and allows for response-guided therapeutic strategies. On the other hand, neo-adjuvant treatment delays resection of the primary tumor and carries the risk of tumor cell dissemination during the treatment interval. In particular for patients who are non-responsive to NaCT, the risk–benefit ratio may be unfavorable. While estrogen receptor (ER)-negative breast tumors show substantial rates of pCR, complete response to NaCT in ER-positive breast tumors is limited.3 Patients with ER-positive breast cancer thus would benefit most from predictive markers that would allow enrichment of patients with potentially responsive disease and avoidance of ineffective treatment for others.
Breast tumor RNA expression profiles have been highly successful as prognostic markers in the post-surgical setting.4,5,6,7,8 A number of RNA expression signatures from formalin-fixed, paraffin-embedded (FFPE) surgical specimens are now commercially available to assess prognosis in early stage, ER-positive breast cancer. Clinical guidelines have incorporated the use of such assays for the purpose of selecting patients with early stage, ER-positive disease for post-surgical chemotherapy.1,9 Each expression score defines patients of low, intermediate, and high risk of recurrence based on the expression of a variable number of genes that consistently include a module measuring proliferation. Several studies have shown that high-risk estimates in breast cancer prognostic profiles are driven primarily by high expression of the proliferation module.10,11
The 21-gene Recurrence Score (RS) and the 70-gene assay (MammaPrint) were the first prognostic assays to achieve clinical acceptance and were validated in prospective randomized trials.12,13 They have been followed by several “second generation” expression signatures [Breast Cancer Index (BCI), 12-gene Molecular Score (MS; EndoPredict), PAM50 risk of recurrence score (Prosigna)].5,6,8 Retrospective analyses in the translational arm of the ATAC trial have allowed direct comparison of four commercial breast cancer prognostic tests, with second generation expression scores generally outperforming the 21-gene RS as prognostic markers.14,15,16
Numerous studies have evaluated individual breast prognostic profiles for their ability to predict response to NaCT and, in some cases, neo-adjuvant endocrine therapy.17,18,19,20,21,22,23,24,25,26 However, little is known of their comparative value in predicting neo-adjuvant response. Here we use six public microarray expression data sets and approximations of one second-generation signature, the 12-gene MS, as well as one first generation signature, the 21-gene RS, to compare their ability to predict response to NaCT from pre-treatment breast tumor biopsies.
Methods
Data Sets
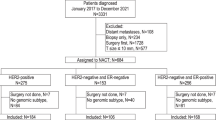
Public microarray gene expression data were obtained from the Gene Expression Omnibus (GEO) database.27 Data sets were selected if patients had been diagnosed with breast cancer, received NaCT, included at least 50 patients with ER-positive, HER2-negative disease, had expression data from pre-treatment fresh-frozen biopsies, made available treatment-associated outcome data on pathological response, and had been analyzed on commercial microarrays. Six data sets met all criteria: GEO accessions GSE20194, GSE20271, GSE25066, GSE32646, GSE34138, and GSE41998.28,29,30,31,32,33 Five studies used Affymetrix Human Genome U133 arrays, one study employed Illumina WG6 v3 bead chips (Table 1). Data from Affymetrix arrays were downloaded as.CEL files and normalized using RMA. Illumina data were between-array normalized by simple scaling (SSN). As all data were from public microarray datasets, oversight by an institutional review board was not required.
Derivation of the 12-Gene Molecular Score
The 12-gene MS contains four normalization and control genes and eight target genes linked to proliferation and the ER pathway. Expression probes for the eight target genes (AZGP1, BIRC5, DHCR7, IL6ST, MGP, RBBP8, STC2, UBE2C) were averaged by gene, and gene averages combined into a 12-gene MS according to the published algorithm.6 At least one probe was available for all genes in all data sets.
Derivation of the 21-Gene Recurrence Score
Sixteen genes in the 21-gene RS are prognostic markers with the remaining five housekeeper genes being used for normalization. Expression probes for the 16 target genes (AURKA, BAG1, BCL2, BIRC5, CCNB1, CD68, CTSL2, ERBB2, ESR1, GRB7, GSTM1, MKI67, MMP11, MYBL2, PGR, SCUBE2) were averaged by gene, and gene averages combined into gene groups for proliferation, ER, HER2 and invasion scores as described.4 At least one probe per gene was available on all arrays with the exception of MYBL2 in GSE34138. A threshold was applied to the proliferation score based on the 80th percentile in ER-positive patients (Supplementary Material).34,35,36 Gene group averages and single gene expression values were combined into a 21-gene RS as published.4
Association with Response
ER-positive, HER2-negative samples were selected based on immunohistochemistry (IHC) or fluorescent in situ hybridization (FISH) status in the accompanying clinical file. For each data set the two expression scores were transformed into z-scores and used as continuous variables. The score ranges differ from those observed on clinical reports from each score due to the different measurement platform and the z-score transformation. Response was defined as pCR obtained from the clinical data. pCR was determined by histological examination of the surgical sample and defined in a similar way across data sets as no residual invasive disease in the breast and lymph nodes. Association of each score with response was tested by logistic regression. To adjust for differences by data set, the cohort name was added as a categorical variable. In the combined analysis the model included both scores and adjustment for cohort. All analysis was performed in R 3.5.0 (R Foundation, 2018).
Results
Of the 1514 patient samples in the six data sets, 782 qualified as ER-positive, HER2-negative by IHC and/or FISH status. Response data were complete for 764 patients with ER-positive, HER2-negative tumors. This included 59 complete pathological responses (8% response rate, Table 1).
Correlation Between Expression Scores
The microarray-approximated 12-gene MS and 21-gene RS were moderately well correlated (r = 0.71) with correlation coefficients similar to those seen in quantitative PCR data.37 Correlations between scores (Supplementary Figure 1) as well as distributions of each score (Supplementary Figure 2) were similar across data sets.
Association with Neo-adjuvant Response
Each expression score was tested for its ability to predict pCR after NaCT in logistic regression models adjusted for cohort. When analyzed separately, either score was predictive of pCR (12-gene MS p = 9.4 × 10−5, 21-gene RS p = 0.0041; Table 2) with higher expression scores indicative of increased probability of response. Probability of response as a function of either expression score is depicted in Fig. 1. The probability of response for the 21-gene RS is higher at low scores and lower at high scores compared to the 12-gene MS; this indicates that the 21-gene RS score does not discriminate probability of response as well as the 12-gene MS. Additionally, when analyzed together in the same model, the 12-gene MS remained a significant predictor of response (p = 0.0079) while the 21-gene RS did not (p = 0.79), indicating that the 12-gene MS has additional discriminatory power not present in the 21-gene RS.
Both the 12-gene MS and the 21-gene RS contain a subset of proliferation genes that are strongly weighted in the respective overall algorithm. However, only the 21-gene RS adds a threshold to the proliferation component.4 To test whether the percentile threshold employed affected the predictive power of the 21-gene RS score we performed a sensitivity analysis. The threshold for the proliferation group was varied from the 75th to the 90th percentile, a range guided by proliferation gene group expression distributions presented in previous publications.34,35,36 The 12-gene MS remained superior to the 21-gene RS in the combined analysis, irrespective of the proliferation group threshold (Supplementary Table 1). While variation in the proliferation threshold did not change the results of the combined analysis, the analyses of the individual score showed increasing loss of discriminatory power of the 21-gene RS with higher proliferation group thresholds.
Discussion
NaCT is an effective tool for down-staging early and locally advanced breast cancer, but the sensitivity to NaCT varies greatly. Clinical parameters such as tumor size and node status have limited utility as response predictors.29,30,38 The most effective marker is ER status. Both in the neo-adjuvant and adjuvant systemic chemotherapy setting, ER-negative breast cancers show better complete response rates and higher absolute chemotherapy benefit, respectively compared to ER-positive tumors.3,39 ER-negative tumors are more highly proliferative and may therefore be particularly sensitive to agents that target DNA replication and/or cell division, such as NaCT. Similarly, luminal B tumors are more proliferative than the luminal A subtype and luminal B tumors have better response rates to NaCT.20 Breast cancer prognostic assays rely in large part on measurement of proliferation gene expression to discriminate risk and response to treatment. It is therefore not surprising that many breast cancer prognostic tests have been shown to predict NaCT.
We have compared the power of two breast cancer prognostic scores to predict NaCT response by using approximations of each score from microarray data. Using probe expression values for the published set of genes we employed the respective algorithms to create a 12-gene MS and 21-gene RS from pre-treatment samples in six NaCT treated breast cancer cohorts. As expected, both scores were predictive of NaCT response, as has been shown in other cohorts.17,21 In direct comparison however, the approximated 12-gene MS was a superior predictor than the approximated 21-gene RS. While the superior prognostic power of the 12-gene MS compared to the 21-gene RS was shown in a head-to-head comparison in the TransATAC cohort,11 this is to our knowledge the first direct comparison of these two prognostic scores as predictive markers for neo NaCT response. The reason for the better predictive power of the 12-gene molecular score could be that this signature includes a strong ER signaling motive beside the proliferation motive. This reflects previous findings that highly proliferative tumors with low ER expression are most sensitive to NaCT.40 A head-to-head comparison in a neo-adjuvant clinical trial could provide further information on the best predictive marker.
Limitations of this study include the use of microarray-based expression data and the limited availability of clinical parameters. The dynamic range for microarray expression data is generally more limited than that for quantitative PCR, especially with the lower end of the expression spectrum being lost to background. However, many of the currently available breast cancer profiles, including the 12-gene MS and the 21-gene RS, were developed from microarray data and the genes selected should be reliably measurable on microarrays.4,6 In addition, expression scores rely heavily on gene sets with generally high expression values in breast tumor tissue, i.e., proliferation and estrogen inducible genes. The 21-gene RS in particular, which thresholds the proliferation gene group upwards of the third quartile of the expression distribution,36 should be minimally affected by lack of discrimination at the lower end of expression values. The correlation between the approximated microarray versions of the 12-gene MS and the 21-gene RS is equivalent to that observed when the two scores are compared using quantitative PCR data.37 This suggests that any effect due to the measurement platform did affect both scores similarly. The observation in the sensitivity analysis of the decreasing power of the 21-gene RS to predict NaCT response with higher proliferation gene group thresholds is intriguing. While the effect of the threshold on the prognostic ability of the 21-gene RS is unknown, it is possible that the better performance of the second-generation prognostic profiles in the TransATAC comparisons may be partially be due to their lack of a proliferation gene threshold, thus enhancing the effect size of proliferation in these algorithms.14,15,16
The lack of availability of extended clinical factors for most public microarray studies prevented the inclusion of clinical parameters such as nodal status and tumor size. However, clinical tumor size and clinical node status are less reliable than post-surgical staging and their utility for predicting NaCT response varies widely across studies.8,29,30,38,41 Tumor features such as receptor status and subtype are more consistent measures of response. Moreover, continuous scores such as the various, widely available prognostic signatures could further improve response prediction. This is particularly desirable for tumor types where new and alternate therapies are available. Accumulating evidence suggests that neo-adjuvant endocrine therapy may be an effective alternate treatment strategy for ER-positive, HER2-negative breast cancer patients.42
Early assessment of tumor response to therapy could limit patient exposure to ineffective treatment and improve tumor control by prioritizing treatment of patients with higher probability of response. Here we show that gene expression scores such as the 12-gene MS and the 21-gene RS are predictive of pCR to NaCT and could therefore inform pre-surgical treatment decisions. Furthermore, since achieving pCR is broadly considered as a surrogate for improved survival in HER2-positive, triple negative, and luminal B breast cancer, both scores might predict long-term benefit from chemotherapy.
References
Gradishar W, Anderson B, Aft R, et al. NCCN clinical practice guidelines in oncology. Breast Cancer. 2018;Version 1.2018. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf. Accessed 23 Aug 2018.
De Mattos-Arruda L, Shen R, Reis-Filho JS, Cortés J. Translating neoadjuvant therapy into survival benefits: one size does not fit all. Nat Rev Clin Oncol. 2016;13:566.
Zambetti M, Mansutti M, Gomez P, et al. Pathological complete response rates following different neoadjuvant chemotherapy regimens for operable breast cancer according to ER status, in two parallel, randomized phase II trials with an adaptive study design (ECTO II). Breast Cancer Res Treat. 2012;132(3):843–51.
Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26.
Nielsen TO, Parker JS, Leung S, et al. A Comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in Tamoxifen-treated estrogen receptor–positive breast cancer. Clin Cancer Res. 2010;16(21):5222.
Filipits M, Rudas M, Jakesz R, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17(18):6012.
Mook S, Schmidt MK, Viale G, et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2009;116(2):295–302.
Zhang Y, Schnabel CA, Schroeder BE, et al. Breast cancer index identifies early-stage estrogen receptor–positive breast cancer patients at risk for early- and late-distant recurrence. Clin Cancer Res. 2013;19(15):4196.
Senkus E, Kyriakides S, Ohno S, et al. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26 Suppl 5:v8–30.
Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genom. 2008;1(1):11.
Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65.
Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–21.
Cardoso F, Vant Veer LJ, Bogaerts J, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375(8):717–29.
Buus R, Sestak I, Kronenwett R, et al. Comparison of EndoPredict and EPclin with Oncotype DX recurrence score for prediction of risk of distant recurrence after endocrine therapy. JNCI J Natl Cancer Inst. 2016;108(11):djw149.
Dowsett M, Sestak I, Lopez-Knowles E, et al. Comparison of PAM50 risk of recurrence score with Oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31(22):2783–90.
Sgroi DC, Sestak I, Cuzick J, et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: a prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 2013;14(11):1067–76.
Bertucci F, Finetti P, Viens P, Birnbaum D. EndoPredict predicts for the response to neoadjuvant chemotherapy in ER-positive, HER2-negative breast cancer. Cancer Lett. 2014;355(1):70–5.
Louis WCC, Satoshi M, Christopher YCC, Wai-Kuen N, Masakazu T. Neoadjuvant palbociclib on ER + breast cancer (N007): clinical response and EndoPredict’s value. Endocr Relat Cancer. 2018;25(2):123–130.
Prat A, Galván P, Jimenez B, et al. Prediction of response to neoadjuvant chemotherapy using core needle biopsy samples with the Prosigna Assay. Clin Cancer Res. 2016;22(3):560.
Prat A, Fan C, Fernández A, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015;13(1):303.
Gianni L, Zambetti M, Clark K, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23(29):7265–77.
Mathieu MC, Mazouni C, Kesty NC, et al. Breast Cancer Index predicts pathological complete response and eligibility for breast conserving surgery in breast cancer patients treated with neoadjuvant chemotherapy. Ann Oncol. 2012;23(8):2046–52.
Straver ME, Glas AM, Hannemann J, et al. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2010;119(3):551–8.
Dunbier AK, Anderson H, Ghazoui Z, et al. Association between breast cancer subtypes and response to neoadjuvant anastrozole. Steroids. 2011;76(8):736–40.
Prat A, Cheang MU, Galván P, et al. Prognostic value of intrinsic subtypes in hormone receptor–positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncol. 2016;2(10):1287–94.
Ueno T, Masuda N, Yamanaka T, et al. Evaluating the 21-gene assay Recurrence Score® as a predictor of clinical response to 24 weeks of neoadjuvant exemestane in estrogen receptor-positive breast cancer. Int J Clin Oncol. 2014;19(4):607–13.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10.
Consortium M, Shi L, Campbell G, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827.
Tabchy A, Valero V, Vidaurre T, et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res. 2010;16(21):5351.
Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–81.
Miyake T, Nakayama T, Naoi Y, et al. GSTP1 expression predicts poor pathological complete response to neoadjuvant chemotherapy in ER-negative breast cancer. Cancer Sci. 2012;103(5):913–20.
de Ronde JJ, Rigaill G, Rottenberg S, Rodenhuis S, Wessels LFA. Identifying subgroup markers in heterogeneous populations. Nucleic Acids Res. 2013;41(21):e200.
Horak CE, Pusztai L, Xing G, et al. Biomarker analysis of neoadjuvant doxorubicin/cyclophosphamide followed by ixabepilone or paclitaxel in early-stage breast vancer. Clin Cancer Res. 2013;19(6):1587.
Anderson JM, Yoshizawa C, Winer EP, et al. Abstract P3-10-15: quantitative gene expression by RT-PCR in classic and variant forms of lobular carcinoma in estrogen receptor positive invasive breast cancer. Cancer Res. 2010;70(24 Supplement):P3-10–15.
Stull TS, Goodwin MC, Anderson JM, et al. P3-06-05: Comparison of Oncotype DX® recurrence scores between surgical and core biopsy specimens in breast cancer patients. Cancer Res. 2011;71(24 Supplement):P3-06–05.
Swain SM, Nunes R, Yoshizawa C, Rothney M, Sing AP. Quantitative gene expression by recurrence score in ER-positive breast cancer, by age. Adv Ther. 2015;32(12):1222–36.
Varga Z, Sinn P, Fritzsche F, et al. Comparison of EndoPredict and Oncotype DX test results in hormone receptor positive invasive breast cancer. PLoS ONE. 2013;8(3):e58483.
Denkert C, Loibl S, Noske A, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28(1):105–13.
Jones RL, Salter J, A’Hern R, et al. Relationship between oestrogen receptor status and proliferation in predicting response and long-term outcome to neoadjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2009;119(2):315.
Landmann A, Farrugia DJ, Zhu L, et al. Low estrogen receptor (ER)–positive breast cancer and neoadjuvant systemic chemotherapy. Is response similar to typical ER-positive or ER-negative disease? Am J Clin Pathol. 2018;150(1):34–42.
Seo AN, Lee HJ, Kim EJ, et al. Tumour-infiltrating CD8 + lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br J Cancer. 2013;109:2705.
Spring LM, Gupta A, Reynolds KL, et al. Neoadjuvant endocrine therapy for estrogen receptor–positive breast cancer: A systematic review and meta-analysis. JAMA Oncol. 2016;2(11):1477–86.
Acknowledgement
The authors would like to acknowledge Stephanie Meek, PhD for her assistance in preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
This study was financially supported by Myriad Genetics, Inc. SW, DF, RK, JML, JSL, and AG were all employed by Myriad Genetics at the time of the study. DF has a patent for a similar technology (PCT/US2015/027091 filed April 22, 2015). MR has served as an advisor for and received travel/accommodation expenses from AstraZeneca; has received institutional research funding from AbbVie, AstraZeneca, Invitae, Medivation, Myriad Genetics, Inc., Pfizer, and Tesaro; has received in-kind research funding from Invitae and Myriad Genetics, Inc.; has received other transfers of value (editorial services) from Pfizer; and his research on this project was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. LS has financial activities with Amgen, GSK, Spectrum, Medivation, Bayer, Genentech, Pfizer, Sanofi, BMS, Novartis, and MedImmune outside of the submitted work. PS has served on an advisory board for Myriad Genetics, Inc. All other authors report no conflicts of interest. This study has previously been presented at ASCO 2018.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Soliman, H., Wagner, S., Flake, D.D. et al. Evaluation of the 12-Gene Molecular Score and the 21-Gene Recurrence Score as Predictors of Response to Neo-adjuvant Chemotherapy in Estrogen Receptor-Positive, HER2-Negative Breast Cancer. Ann Surg Oncol 27, 765–771 (2020). https://doi.org/10.1245/s10434-019-08039-7
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-019-08039-7