Abstract
Stepwise covariate modeling (SCM) has a high computational burden and can select the wrong covariates. Machine learning (ML) has been proposed as a screening tool to improve the efficiency of covariate selection, but little is known about how to apply ML on actual clinical data. First, we simulated datasets based on clinical data to compare the performance of various ML and traditional pharmacometrics (PMX) techniques with and without accounting for highly-correlated covariates. This simulation step identified the ML algorithm and the number of top covariates to select when using the actual clinical data. A previously developed desipramine population-pharmacokinetic model was used to simulate virtual subjects. Fifteen covariates were considered with four having an effect included. Based on the F1 score (an accuracy measure), ridge regression was the most accurate ML technique on 200 simulated datasets (F1 score = 0.475 ± 0.231), a performance which almost doubled when highly-correlated covariates were accounted for (F1 score = 0.860 ± 0.158). These performances were better than forwards selection with SCM (F1 score = 0.251 ± 0.274 and 0.499 ± 0.381 without/with correlations respectively). In terms of computational cost, ridge regression (0.42 ± 0.07 seconds/simulated dataset, 1 thread) was ~20,000 times faster than SCM (2.30 ± 2.29 hours, 15 threads). On the clinical dataset, prescreening with the selected ML algorithm reduced SCM runtime by 42.86% (from 1.75 to 1.00 days) and produced the same final model as SCM only. In conclusion, we have demonstrated that accounting for highly-correlated covariates improves ML prescreening accuracy. The choice of ML method and the proportion of important covariates (unknown a priori) can be guided by simulations.
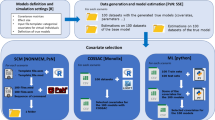
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Covariate modeling is an important aspect of population pharmacokinetics and can influence key drug development decisions such as dose optimization in patient subpopulations and identification of subpopulation key features (1, 2). Stepwise covariate modeling (SCM) is the most commonly used technique for covariate selection (1, 3). However, it has a high computational burden especially for complex structural models or a high number of candidate covariates (1, 4). Increasing model complexity and too many false highly-correlated covariates also decrease its power to detect the true covariates (5). Moreover, the emerging role of quantitative systems pharmacology and real-world data in informing clinical drug development (6,7,8), means that pharmacometrics (PMX) models are becoming more complex and the number of candidate covariates keeps increasing.
To increase computational speed, one solution is to use first-order conditional estimation (FOCE)-based linear approximation (FOCE linearization), previously demonstrated to reduce runtimes from days to less than 10 minutes (9). Another common solution is to screen covariates prior to covariate modeling using techniques such as graphical analysis (e.g. empirical Bayes estimates [EBEs] of a parameter versus covariates) or statistical techniques such as correlation analysis, and regression-based methods (1, 2). These methods are often applied to low dimensional problems (i.e., problems involving a relatively small number of covariates) and are often based on prior knowledge, which can sometimes be biased (4).
Supervised machine learning (ML), which utilizes statistical techniques to deal with both low-and multi-dimensional problems, has been proposed as a covariate screening tool (4, 10, 11). For example, Sibieude and colleagues used different scenarios of covariate influence in simulated datasets to compare three ML methods (random forest, neural networks, and support vector regression) with traditional PMX approaches (i.e., SCM, conditional sampling use for stepwise approach based on correlation tests [COSSAC] and least absolute shrinkage and selection operator [LASSO]) and concluded that ML methods can increase the efficiency of covariate modeling (similar or better performance than SCM with 30 to 100 times decreased computational time burden) (4). They recommended that more conventional PMX approaches be performed after the initial covariate screening to obtain the covariate relationships and effects for the final model. Sibieude et al. used a one-compartment model, and did not account for highly-correlated covariates; given that both model complexity and high correlations affect covariate selection accuracy (5), more work in this area is required. Particularly, machine learning utilizes empirical Bayes estimates [EBEs] and whether EBEs effectively retain sufficient information to identify true covariates for mechanistically more complex models as they did for the simpler model will remain an area of active ML research in the near future. Accounting for correlations is especially important during screening to avoid excluding physiologically-relevant covariates due to the selection of highly-correlated false covariates.
Desipramine, a commonly used antidepressant, is extensively metabolized by cytochrome P450 family 2 subfamily D member 6 (CYP2D6), and is listed by the FDA and EMA as a clinical probe substrate for the evaluation of CYP2D6-mediated drug-drug interactions (DDIs) (12, 13). Following oral administration, desipramine is rapidly absorbed with its maximum plasma concentration (Cmax) being observed within 2 to 6 hours (12). It is highly tissue-bound (apparent volume of distribution ranging from 10 to 50 Lkg−1), highly protein-bound (range 73 to 92%), and its bioavailability (~40%) is highly variable (14). Desipramine is a moderate extraction ratio drug mainly eliminated by hepatic metabolism with its main metabolite being 2-OH-desipramine (12, 14).
Previously, Gueorguieva and colleagues used data from control groups (desipramine-only arm) of seven DDI studies (108 healthy subjects) to, among other aims, develop a population pharmacokinetic (pop-PK) model with hepatic intrinsic clearance derived from the total clearance (12). During covariate modeling, demographic characteristics (age, alcohol status, body mass index [BMI], gender, height, race, weight, and smoking status) were tested on apparent clearance and volume of distribution parameters using SCM, with only BMI being included as a significant covariate on the volume of distribution (12).
In earlier analysis, using a slightly larger sample size (114 healthy subjects) and additional covariates, we built upon Gueorguieva et al.’s work and developed a semi-mechanistic pop-PK structural model as well as determined the influence of including below limit of quantification data (unpublished data, Ndzamba, Asiimwe, Mouksassi, Pillai, Lombard, and Lang). This paper focuses on examining the role of machine learning for covariate screening prior to covariate selection. Specifically, we a) compared the performance of various ML and PMX techniques on datasets simulated using a more complex model structure and clinical trial data; b) explored whether accounting for highly-correlated covariates improves the true covariate selection accuracy; and, c) applied the best-performing ML techniques to clinical trial data.
Materials and Methods
To select the machine learning (ML) technique to apply to the desipramine dataset, we conducted a simulation analysis and compared the performances of the various ML techniques. Based on the framework proposed by Sibieude et al. (4), our analysis workflow (Fig. 1) included structural model definition, simulation settings, data generation, and model estimation before covariate selection, where we compared the various ML techniques, and included the traditional pharmacometrics (PMX) techniques. The different components of the analysis workflow are described in more details below (relevant code is provided in Supplementary Code).
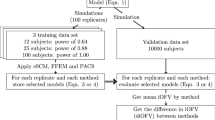
Analysis (simulations and performances of the various ML techniques) workflow. aThe large effect size was 1.5 for the categorical covariate and 1 for the continuous covariates while the small effect size was 1.15 for the categorical covariate and 0.15 for the continuous covariates. AIC = Akaike information criterion, EBEs = empirical bayes estimates, FOCE = first-order conditional estimation, GAM = generalized additive model (using splines), MSE = mean squared error, NONMEM = NONlinear Mixed Effects Modeling software, PMX = pharmacometrics, PsN = perl speaks NONMEM, RMSE = root mean squared error, SCM = stepwise covariate modeling
Desipramine Dataset
The desipramine dataset included 114 subjects recruited from seven DDI studies whose selected characteristics are shown in Table S1 (12). It is worth noting that although machine learning could have handled all available 88 covariates (collected by at least one of the seven studies) or 33 covariates (available in all studies), we selected the most relevant 15 (Table S1, Fig. S1) to optimize the runtime for SCM that did not have ML prescreening.
Structural Model
We used a semi-mechanistic first-order absorption model with a liver compartment, a central compartment, a peripheral compartment and 3 transit compartments. This model (scheme in Fig. S2 and parameter estimates in Table I) was developed from our earlier analysis (unpublished data, Ndzamba, Asiimwe, Mouksassi, Pillai, Lombard, and Lang) using nonlinear mixed-effects modeling in NONMEM (version 7.5.0/7.5.1) with Perl Speaks NONMEM (PsN, version 5.3.0) (15).
Settings and Simulation Framework of 200 A Priori ‘True’ Models
To select the appropriate ML model to apply to the clinical dataset, we defined a simulation framework. We generated 200 simulated datasets (100 per scenario, see below) using the structural model and assumed a priori 4 true covariates (one ‘true’ categorical covariate [formulation] and three ‘true’ continuous covariates). The fifteen covariates from the clinical dataset shown in Table S1 were included in (used to inform) the simulation analysis as detailed in Text S1. We defined the a priori ‘true’ covariate effects on the intrinsic hepatic clearance (CLint) parameter to derive the ‘true’ model, using power and linear relationships for continuous and categorical covariates, respectively.
We included three a priori ‘true’ continuous covariates based on the number of high correlations (defined as correlation coefficients >0.5) with other continuous covariates: age (zero correlation with continuous covariates), fat-free mass FFM (highly-correlated with one continuous covariate [weight]), and aspartate aminotransferase AST (highly-correlated with three continuous covariates [alanine aminotransferase ALT, bilirubin and gamma-glutamyl transferase GGT]) (Fig. S3).
We explored two scenarios: a ‘large effect’ scenario in which [the non-reference] formulation had an effect size of 1.5 (or 50% higher CLint with a linear relationship) and the continuous covariates an effect size of 1 (see Fig. S4 for interpretation); and a ‘small effect’ scenario in which formulation had an effect size of 1.15 (or 15% higher CLint) and the continuous covariates an effect size of 0.15 (Fig. S4 for details).
To derive the a priori ‘true’ model, the parameter estimates and covariance matrix for random effects/ETAs of the structural model including the a priori ‘true’ covariate effects were used to simulate 100 datasets per scenario in R (version 4.3.1) (16) (package rxode2 (17)). Each dataset comprised of 114 virtual subjects (similar to the desipramine dataset) with fifteen sampling times (0, 0.5, 1, 2, 3, 4, 6, 8, 12, 24, 48, 72, 96, 120, 144 hours).
Finally, the a priori ‘true’ model was estimated for each scenario/simulated dataset in NONMEM. The empirical Bayes estimates (EBEs) were then used as outcomes in the ML analysis.
Evaluation of ML and PMX Techniques for Covariate Selection for the A Priori ‘True’ Models
The traditional PMX techniques that we tested were: [non-linear] SCM (forward addition with and without backward selection) and SCM with FOCE linearization (9). SCM options included: forward p value of 0.05, backwards p value of 0.01, linear relationships for categorical covariates and linear/piecewise/exponential/power relationships for continuous covariates.
For machine learning, we used the Caret package in R (18) to implement several techniques including linear regression and generalized additive model using splines (fast, simple and commonly used), penalized regression (penalty terms are included on the model’s objective functions; includes LASSO [alpha = 1], ridge regression [alpha = 0] and elastic net [0 < alpha <1]), step-wise linear methods (based on Akaike information criterion/AIC and root mean squared error/RMSE; similar in principle to SCM), random forest (an ensemble learning method that combines the predictions of multiple decision trees to make a final decision), neural networks (inspired by the structure of biological neural networks and can learn complex patterns and relationships in the data) and extreme gradient boosting/XGBoost (another ensemble method that builds a sequence of models, each one improving the predictions of the previous model). Additional information about these machine learning techniques is provided in Table S2. Tuning of hyperparameters (e.g., alpha for penalized regression or number of trees for tree-based methods such as random forest) was achieved in Caret using five-fold cross-validation and default settings (see Table S2 for the parameters that were tuned).
We first used the NONMEM CLint individual random effects/ETAs for the ML analysis, where they were analyzed as the outcome. To compare the use of individual THETA parameters instead of ETAs, we also carried out additional analysis using logarithmic transformed THETAs. For consistency with traditional PMX techniques, all continuous covariates were scaled by dividing them using their median value (or 70 kg for weight) followed by a logarithmic transformation (equivalent to a power relation on the linear scale). For some ML methods (penalized regression and neural networks), an additional transformation (minimum-maximum normalization) was done.
To rank the most important covariates in each ML technique, variable importance was computed either using model-based metrics (e.g., t-statistic absolute value for linear models, out-of-bag mean squared error for random forest) or model-independent metrics (e.g., R-squared statistic, absolute value of the t-value for the slope of the linear predictor) (18). We then selected the top-4 covariates (top-M approach), where M is the number of true covariates (4). From this, the F1-score (the harmonic mean of recall [true positive/(true positive + false negative)] and precision [true positive/(true positive + false positive)]) was calculated (F1 score = 2 x (recall x precision)/(recall + precision)) and used to compare the ML and PMX results (4).
Accounting for Highly-Correlated Covariates to Determine the Important Covariates
Because a highly-correlated covariate can be selected instead of the ‘true’ covariate (19), we also computed the F1 score by accounting for highly-correlated covariates, defined as a correlation coefficient > 0.5 for relationships involving only continuous covariates or a Fisher’s exact test/Mann-Whitney p value <0.05 for relationships involving categorical covariates. When accounting for highly-correlated covariates, a true covariate was considered to have been selected if it or any of its highly-correlated covariates was selected.
Application of the Best ML Methods to the Desipramine/Clinical Trial Dataset
The best ML methods identified during the simulation analysis were applied to the desipramine dataset to screen 15 covariates (Table S1) on three parameters (CLint, transit rate constant/KTR and apparent central volume of distribution/Vc; Table S3) prior to non-linear SCM (linear relationships for categorical covariates and linear/power relationships for continuous covariates). This was compared to non-linear SCM on the 15 covariates without ML screening. The ML methods were also applied to a larger set of covariates (n = 33) but, no comparison was made with SCM because of very high computational requirements. As the number of true covariates was unknown a priori on the desipramine dataset, the top-M approach (where M = number of true covariates) was not applicable. We therefore determined the number of covariates to test based on the results of the simulation analysis. Specifically, M was set to the median proportion of covariates selected by an ML technique during the simulation analysis.
Results
Machine Learning Improves the Speed and Accuracy of Covariate Selection
We first used individual ETAs on intrinsic hepatic clearance as the outcome, with the ETA shrinkage being <0.1% in all simulated datasets. Without accounting for highly-correlated covariates, ridge regression (F1 score for both scenarios = 0.475 ± 0.231) was the best ML technique (Table II). The PMX techniques performed worse than the ML methods with non-linear SCM (standard approach) having an F1 score of 0.251 ± 0.274 with only forwards selection, and 0.206 ± 0.228 with both forwards and backwards selection. In terms of perfect selection (i.e., number of simulated datasets in which the F1 score was 1, or in which only the true covariates were identified), all methods performed poorly as detailed in Table II. Because smaller covariate effects are harder to detect, all the F1 scores decreased in the ‘low effect’ scenario (F1 scores for ridge regression were 0.590 and 0.360 in the high and low effect scenarios, respectively). Using log-transformed individual THETAs as outcome for the ML techniques (Table S4) produced results that were similar to using the ETAs. The number of times the a priori ‘true’ covariates were included for each of the selection methods are shown in Fig. 2 (panel A), with ridge regression selecting formulation, AST, age, and FFM in 21.0%, 55.0%, 61.5% and 52.5% of the 200 simulated datasets (both scenarios).
True covariates selected by the different methods in both the high and low effect scenarios (200 simulated datasets) during the simulation analysis, with methods ranked according to the F1-score. Panels a and b respectively show covariate selections without and with adjustment for highly-correlated covariates while panel c shows the differences in selection between panels a and b for each of the methods. NONMEM fixed effects/ETAs were used as outcome for the ML methods. AIC = Akaike information criteria, ALT = alanine aminotransferase, AST = aspartate aminotransferase, BMI = body-mass index, BS = backwards selection, FFM = fat-free mass, FS = forwards selection, GAM = generalized additive model (using splines), GGT = gamma-glutamyl transferase, LASSO = least absolute shrinkage and selection operator, ML = machine learning, PMX = pharmacometrics, reg = regression, RMSE = root mean squared error, SCM = stepwise covariate modeling, SCML = stepwise covariate modeling with first-order conditional estimation-based linearization, XGBoost = extreme gradient boosting
The computational cost per simulated dataset for the various methods is shown in Table S5. The fastest ML technique was RMSE-based stepwise selection with a mean (± SD) runtime of 0.25 ± 0.03 seconds in the analysis that used NONMEM ETAs. The most accurate technique (ridge regression) had a runtime of 0.42 ± 0.07 seconds. The PMX methods were much slower than the ML techniques with FOCE-linearized SCM (both forwards and backwards selection) using 15 threads requiring 544.56 ± 99.71 seconds (9.08 ± 1.66 minutes) and non-linear SCM (both forwards and backwards selection, 15 threads) requiring 8288.17 ± 8249.39 seconds (2.30 ± 2.29 hours). This means that ridge regression (using one thread) was ~20,000 times faster than the commonly used non-linear SCM (15 threads).
Accounting for Highly-Correlated Covariates Increases True Covariate Selection Accuracy
When we accounted for correlations (i.e., assumed that including a highly-correlated covariate was comparable to including the true covariate; Table III), ridge regression (F1 score for both scenarios = 0.860 ± 0.158) was still the best technique. The PMX techniques again performed worse than the ML methods with non-linear SCM having an F1 score of 0.499 ± 0.381 with only forwards selection, and 0.444 ± 0.357 with both forwards and backwards selection. For all methods, accounting/adjusting for highly-correlated covariates (Table III) approximately increased F1 scores by two-fold compared to those obtained when no correlations were considered (Table II). Perfect selection scores (i.e., number of simulated datasets in which the F1 score was 1, or in which only the ‘true’ covariates or their correlations were identified) significantly increased, with elastic net regression (113/200 datasets with F1 score = 1) having the best performance. Again, using log-transformed individual THETAs as outcome for the ML techniques produced results (Table S6) similar to using the ETAs.
Figure 2 (panels A-without and B-with correlations) shows that accounting for correlations increased true covariate selection (panel C); for ridge regression, this increased by 10.0% for age (61.5% vs 71.5%), 26.5% for FFM (52.5% vs 79.0%), 38.5% for AST (55.0% vs 93.5%) and 79.0% for formulation (21.0% vs 100.0%). These increments were consistent with the median [range] number of correlations in the simulated datasets (age: 1 [1 to 3], FFM: 2 [1 to 5], AST: 4 [4 to 6], and formulation: 7 [5 to 10]). Covariate selection patterns for both scenarios are shown in Fig. 3 (ridge regression) and Fig. S5 (all methods). As shown for ridge regression, covariates that were correlated with the ‘true’ covariates were almost always selected together with the ‘true’ covariate. Specifically, the three covariates (bilirubin, ALT and GGT) that were correlated to AST (Fig. S3) had a similar selection frequency ranging from 93.5% to 94.5%. Lastly, Fig. S6 shows how the different methods ranked all the 15 covariates.
Covariate ranking and selection pattern for ridge regression in both the high and low effect scenarios (200 simulated datasets) during the simulation analysis. Panel a shows how the four true covariates were ranked across the different methods. The covariate selection frequencies for ridge regression are annotated, with the most ranked covariate (formulation, selected in 100% of the datasets) appearing on top. Panels b (correlations not accounted for) and c (correlations accounted for) show the frequency of selection of all covariates including the true covariates shown in Fig. 2. For ridge regression, accounting for correlations increased the covariate selection frequency by 10.0%, 26.5%, 38.5%, and 79.0% for the four true covariates age, FFM, AST, and formulation respectively. Panel d shows how often covariates are selected together while panel e shows which covariates are selected together. For instance, the first column in both panels shows that formulation, ALT, bilirubin, AST, GGT, weight, FFM, age and BMI were selected together in 21 datasets. Looking at the entire panel e shows that some covariates, specifically ALT, bilirubin, GGT, and AST were almost always selected together, an observation which can be explained by their being highly correlated (correlation coefficient > 0.5). NONMEM ETAs were used as outcome for the ML methods, and except for panel b, correlations between covariates were accounted for. AIC = Akaike information criteria, ALT = alanine aminotransferase, AST = aspartate aminotransferase, BMI = body-mass index, BS = backwards selection, FFM = fat-free mass, FS = forwards selection, GAM = generalized additive model (using splines), GGT = gamma-glutamyl transferase, LASSO = least absolute shrinkage and selection operator, RMSE = root mean squared error, SCM = stepwise covariate modeling, SCML = stepwise covariate modeling with first-order conditional estimation-based linearization, XGBoost = extreme gradient boosting
Simulations Can Inform the Application of Machine Learning to Clinical Trial Data
On the desipramine dataset, we applied both non-linear SCM and ridge/elastic net regression (Tables S7 and S8) followed by non-linear SCM. In the 200 simulated datasets (Table III), ridge and elastic net regression selected a median of 9 covariates (range 2 to 13), which means they decreased the number of candidate covariates by an average of 40%. Applying this figure to the results in Tables S7 and S8 means we tested nine covariates (AST, ALT, bilirubin, estimated glomerular filtration rate/eGFR, formulation, GGT, race, sex, and smoking status) on the CLint, five covariates (age, alcohol status, formulation, race and sex) on the KTR, and eight covariates (ALT, AST, BMI, FFM, formulation, GGT, sex, and weight) on the Vc parameters. The same final model (unpublished data, Ndzamba, Asiimwe, Mouksassi, Pillai, Lombard, and Lang) with formulation (on all three parameters), FFM (Vc) and bilirubin and race (CLint) was obtained during SCM, with and without penalized-regression preselection. Due to fewer covariates, preselection with penalized regression reduced SCM runtime (6 threads) by 42.86% (from 1.75 to 1.00 days).
The results of applying ML on the desipramine dataset comprising 33 covariates are shown in Tables S9 and S10. A simulation analysis to estimate M using a similar model structure (i.e. four a priori ‘true’ covariates on CLint) put the estimate at 8 (range 5 to 15) for both ridge (attained an F1 score of 0.562 ± 0.277 with correlations accounted for) and elastic net regression (F1 score = 0.566 ± 0.303), which means that starting with 33 candidate covariates and applying machine learning would reduce the number of candidate covariates by an average of 76%. Importantly, this simulation analysis took ~6.1 minutes (applying ridge and elastic net regression on the desipramine dataset took about two and ten seconds, respectively).
Discussion
To examine the role of machine learning (ML) for covariate screening, we used a semi-mechanistic first order absorption structural model with a liver compartment, a central compartment, a peripheral compartment and 3 transit compartments to simulate pharmacokinetic profiles of 114 virtual subjects. Two hundred datasets were simulated and used to test several ML and pharmacometrics (PMX) methods for their ability to select true covariates. As previously reported by Sibieude et al. (4), the ML methods performed better than the PMX methods (e.g., without accounting for highly-correlated covariates, best ML method F1 score = 0.475 vs best PMX method F1 score = 0.251). In Sibieude et al.’s analysis, neural networks had the highest F1 score (0.89 ± 0.16), a score almost twice that achieved by our best ML method. This, however, is unsurprising since Sibieude et al. used a simpler (one-compartment) model and their F1 scores included results from scenarios with few highly-correlated covariates; both model complexity and many false highly-correlated covariates decrease covariate selection accuracy (5). Whereas the overall performance for the ML methods was similar in Sibieude et al.’s analysis (F1 scores = 0.85 to 0.89), in our analysis, ridge regression outperformed some of the other techniques. During model-fitting, we used five-fold cross-validation that was automated through the R caret package (18). This simplified the analysis but a relatively narrow search grid may have sub-optimally tuned some ML methods leading to lower performance. In terms of computational cost and consistent with the use of empirical Bayes estimates/EBEs (i.e. no need to re-estimate the pharmacokinetic models), ML methods performed better than PMX methods (ridge regression using one thread was ~20,000 times faster than non-linear SCM on 15 threads), which is consistent with Sibieude et al.’s study (4). For non-linear SCM and due to a significantly higher computational cost, we were unable to complete analysis using one thread, which emphasizes the importance of ML methods, especially in low-income settings, where modelers may not have access to powerful computers or clusters that can handle multi-threading or parallel computing. FOCE-linearization improved SCM’s runtime (from ~2.30 hours to 9.08 minutes), although this decreased accuracy. The SCM runtimes include the backwards selection step and testing of multiple relations (linear/piecewise/exponential/power) for continuous covariates; excluding backwards selection and testing only power relations for the continuous covariates can reduce runtime, but this would still be much longer than that of the most efficient ML techniques. A p value more stringent than the 0.05 that we used during the forward step could also have reduced runtime, but this would have reduced accuracy (4).
Despite poorer performance with the simulated datasets, we relied on SCM during covariate selection for the desipramine/clinical trial dataset, as it remains the gold standard of covariate modeling. However, it can select the wrong covariates (2, 5), which emphasizes the need for more accurate covariate modeling approaches. If ML can be used to accurately screen covariates to a manageable number, then expert opinion to determine clinical relevance, or evaluation of all possible combinations of covariates (20, 21) can be done to improve the accuracy of covariate selection. With regard to including false highly-correlated covariates instead of the true covariate, this leads to comparable results in terms of model predictions but can lead to interpretation difficulties for which pharmacological understanding would be crucial for any decision making (19). As our results show, accounting for correlations leads to increased accuracy and increased the percentage selection of the four true covariates by 10% (age), 26.5% (FFM), 38.5% (AST) and 79% (formulation) for ridge regression, increments that are consistent with the median number of correlations in the simulated datasets (age: 1, FFM: 2, AST: 4, and formulation: 7). As Fig. 2 shows, some ML techniques may be more sensitive to highly-correlated covariates, with extreme gradient boosting picking up formulation in 0/200 simulated datasets without adjustment for correlation (versus 200/200 simulated datasets with correlation). For the least correlated covariate (age), extreme gradient boosting (and random forest) saw no improvement in covariate selection accuracy when correlations were accounted for. The downside of including correlations is a higher number of retained covariates. During screening, retaining very few covariates should not compromise accuracy, and although we did not do it in our analysis, it is possible to test several thresholds of the correlation coefficient/p-values to choose those that minimize the number of retained covariates while maximizing the F1 score/accuracy.
Ridge regression (highest F1 score) and elastic net regression (highest number of simulated datasets with F1 score = 1) were applied to the desipramine dataset, and preselection with these methods reduced SCM runtime by 42.9%. When applying these methods to the desipramine dataset, we relied on the results of the simulation analysis (these methods selected ~60% of the top covariates) as we could not use the top-M approach, where M is the number of true covariates (unknown a priori). The simulation analysis was conducted using CLint, and so the 60% may have been inappropriate for the KTR and Vc parameters. We also included four a priori true covariates in our simulation analysis, and it is possible that we would have obtained a different percentage had we defined another number of true covariates. Another approach would involve a modeler reviewing the ML output and selecting a given number of top covariates with their correlations. In this study, selecting an arbitrary top three/four covariates with their correlations would produce a similar number of covariates to selecting the top 60% covariates as done during the simulation-based approach. This approach also demonstrates the facilitating role of ML in covariate selection, i.e., that decision-making ultimately rests with the modeler. Although this wasn’t done, combining a ML prescreening followed by a physiologically-relevant screening is recommended and would have saved more SCM run-time. For example, rather than test nine covariates on the CLint (all, except race, were highly correlated), we could have selected one/two most physiologically-relevant (of the highly-correlated) covariates and tested it/them with race. Additionally, an experienced modeler can add back covariates known to be important that have been omitted during ML prescreening. There are several other approaches, such as selecting all covariates assigned an importance score above zero for selection methods such as LASSO and elastic-net or using techniques such as BORUTA (22) that can employ shadow variables (permuted copies of the original variables) to decide which variables are important. Covariate selection with ML is still an emerging field and therefore further study is needed to determine the best approaches that can be implemented a priori. Another area that will benefit from further study is how to account for correlations between the parameters. In the current study, the NONMEM EBEs were evaluated independently for the three parameters when applying the ML methods to the desipramine dataset. SCM can account for these correlations and when CLint was tested alone in a post-hoc analysis, the selected covariates were fat-free mass and formulation (when tested together with KTR and Vc, bilirubin, formulation, and race were selected on CLint).
Our study had challenges additional to a relatively narrow search grid during parameter tuning and data limitations (e.g., inability to explore a larger range of CYP2D6 metabolizer phenotypes [only normal/extensive metabolizers were recruited], unpublished data, Ndzamba, Asiimwe, Mouksassi, Pillai, Lombard, and Lang). For instance, although we tested SCM with FOCE linearization, we did not test other implementations that could have increased speed including SCM+, that can reduce the number of executed models by up to 70% (1). However, SCM+ is not as efficient as FOCE linearization (1, 9) that we tested. We also limited the number of tested covariates to 15 to be able to compare machine learning with the computationally expensive non-linear SCM. This means that when we analyzed 33 covariates, we could not compare our results with SCM. Indeed, given that step-wise methods ignore portions of the model space (23), they are highly-sensitive to the starting set of covariates and comparison with SCM with 15 covariates would not be appropriate. The analysis involving 33 covariates also used only two ML techniques, that both, as expected due to more correlations (5), had decreased performance during the simulation analysis. Consequently, we cannot ascertain if other ML methods might have performed better. However, given that the PMX techniques couldn’t be explored, adding additional ML techniques wouldn’t have added value when comparing ML with PMX methods. In the analysis involving 15 covariates, we selected the ML techniques, that in our opinion, were of main interest. This means that we may have missed out on superior techniques, including ensembles that include a diverse set of ML methods (e.g., LASSO followed by neural networks or tree-based methods). However, including these techniques would not change the conclusions that we made with the ML methods used i.e., that ML may be better than PMX methods. Also, the sample size used during the ML analysis may have been insufficient for some techniques. Penalized (including ridge and elastic net) regression is able to handle multicollinearity with small sample sizes (24), and this could have contributed to its superior performance. However, techniques such as neural networks or random forest are data-hungry and for a continuous outcome like the NONMEM EBEs, may require a subject-per-candidate covariate parameter in excess of 200 (25), or for the 15 evaluated covariates, a minimum sample size of 3000 subjects. A relatively small sample size also means that cross-validation may produce biased estimates and hyperparameter tuning may be more sensitive to the randomization seed used (affecting the test-train split), which would in turn impact the performance of sensitive methods (26, 27). By trying different randomization seeds, the F1 scores of the top penalized regression methods remained similar which showed stability and less dependence on the hyperparameter settings. Lastly, we did not explore time-varying covariates as this was outside the scope of the current analysis.
Conclusion
In conclusion, although the ML methods still need further evaluation for more complex models, different and larger datasets and our conclusion may not be generally applicable for other cases, we have demonstrated using a semi-mechanistic desipramine pop-PK model that ML methods can improve the efficiency of covariate selection by facilitating a speedy covariate screening process. In our dataset that had a moderate number of subjects (n = 114) and many highly-correlated covariates, penalized regression methods performed better than other methods during covariate prescreening. Given the established benefits of penalized regression in addressing multicollinearity and its capacity to handle high-dimensional datasets (28), we believe that penalized regression can effectively perform in high-dimensional settings that encompass many covariates, including highly correlated ones, regardless of the model structure. Using ML methods during covariate prescreening is becoming increasingly relevant as studies are now collecting more and more data (4), including genomic data that can include millions of genetic variants and for which prescreening based on graphical analysis and prior knowledge is not possible. Moreover, computational gain is more pronounced in more complex structural models as the performance of ML is not influenced by model complexity since it does not involve solving the ordinary differential equations (4). We have also demonstrated that accounting for highly-correlated covariates during ML prescreening is crucial to reducing the likelihood of omitting true/physiologically-relevant covariates. Applying ML prescreening to clinical trial data requires some knowledge of the true covariates, which are however unknown a priori; in this work, we also demonstrated how simulations (based on the clinical trial data) can be used to decide which ML technique is most likely to accurately select covariates given the available dataset as well as guide the proportion of important covariate selection. After covariate screening, model building can follow the traditional PMX approaches, such as selection of the most physiologically-relevant covariates by the modeler, followed by SCM and model assessment through graphical checks, clinical relevance etc. Importantly, any data analyst who uses ML approaches should carefully assess the methods for their specific purpose and suitability to their datasets. Improving the efficiency of covariate selection through an ML-facilitated covariate prescreening process could lead to quicker decision-making during drug discovery and development.
Data Availability
The data that support the findings of this study are available from Eli Lilly & Company but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. The code necessary to reproduce the datasets used during the simulation analyses is included in this published article and its supplementary information files.
References
Svensson RJ, Jonsson EN. Efficient and relevant stepwise covariate model building for pharmacometrics. CPT Pharmacometrics Syst Pharmacol. 2022;11(9):1210–22.
Mould DR, Upton RN. Basic concepts in population modeling, simulation, and model-based drug development-part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacometrics Syst Pharmacol. 2013;2(4):e38.
Jonsson EN, Karlsson MO. Automated covariate model building within NONMEM. Pharm Res. 1998;15(9):1463–8.
Sibieude E, Khandelwal A, Hesthaven JS, Girard P, Terranova N. Fast screening of covariates in population models empowered by machine learning. J Pharmacokinet Pharmacodyn. 2021;48(4):597–609.
Ahamadi M, Largajolli A, Diderichsen PM, de Greef R, Kerbusch T, Witjes H, et al. Operating characteristics of stepwise covariate selection in pharmacometric modeling. J Pharmacokinet Pharmacodyn. 2019;46(3):273–85.
Zhao X, Iqbal S, Valdes IL, Dresser M, Girish S. Integrating real-world data to accelerate and guide drug development: a clinical pharmacology perspective. Clin Transl Sci. 2022;15(10):2293–302.
Zhu R, Vora B, Menon S, Younis I, Dwivedi G, Meng Z, et al. Clinical pharmacology applications of real-world data and real-world evidence in drug development and approval-an industry perspective. Clin Pharmacol Ther. 2023;
Mentre F, Friberg LE, Duffull S, French J, Lauffenburger DA, Li L, et al. Pharmacometrics and systems pharmacology 2030. Clin Pharmacol Ther. 2020;107(1):76–8.
Khandelwal A, Harling K, Jonsson EN, Hooker AC, Karlsson MO. A fast method for testing covariates in population PK/PD models. AAPS J. 2011;13(3):464–72.
Sibieude E, Khandelwal A, Girard P, Hesthaven JS, Terranova N. Population pharmacokinetic model selection assisted by machine learning. J Pharmacokinet Pharmacodyn. 2022;49(2):257–70.
McComb M, Bies R, Ramanathan M. Machine learning in pharmacometrics: opportunities and challenges. Br J Clin Pharmacol. 2022;88(4):1482–99.
Gueorguieva I, Jackson K, Wrighton SA, Sinha VP, Chien JY. Desipramine, substrate for CYP2D6 activity: population pharmacokinetic model and design elements of drug-drug interaction trials. Br J Clin Pharmacol. 2010;70(4):523–36.
European Medicines Agency. Guideline on the investigation of drug interactions. In: (CHMP) CfHMP, editor. 2015. p. 1-59.
Ciraulo DA, Barnhill JG, Jaffe JH. Clinical pharmacokinetics of imipramine and desipramine in alcoholics and normal volunteers. Clin Pharmacol Ther. 1988;43(5):509–18.
Keizer RJ, Karlsson MO, Hooker A. Modeling and simulation workbench for NONMEM: tutorial on Pirana, PsN, and Xpose. CPT Pharmacometrics Syst Pharmacol. 2013;2(6):e50.
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 4.2.1 ed. Vienna, Austria2022.
Wang W, Hallow KM, James DA. A tutorial on RxODE: simulating differential equation Pharmacometric models in R. CPT Pharmacometrics Syst Pharmacol. 2016;5(1):3–10.
Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28(5):1–26.
Amann LF, Wicha SG. Operational characteristics of full random effects modelling ('frem') compared to stepwise covariate modelling ('scm'). J Pharmacokinet Pharmacodyn. 2023;
Schatzoff M, Tsao R, Fienberg S. Efficient calculation of all possible regressions. Technometrics. 1968;10(4):769–79.
Heinze G, Wallisch C, Dunkler D. Variable selection - a review and recommendations for the practicing statistician. Biom J. 2018;60(3):431–49.
Kursa MB, Rudnicki WR. Feature selection with the Boruta package. J Stat Softw. 2010;36(11):1–13.
Hutmacher MM, Kowalski KG. Covariate selection in pharmacometric analyses: a review of methods. Br J Clin Pharmacol. 2015;79(1):132–47.
Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc Ser B Methodol. 1996;58(1):267–88.
van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. 2014;14:137.
Vabalas A, Gowen E, Poliakoff E, Casson AJ. Machine learning algorithm validation with a limited sample size. PLoS One. 2019;14(11):e0224365.
Song L, Minku L, Yao X, editors. The impact of parameter tuning on software effort estimation using learning machines. Proceedings of the 9th International Conference on Predictive Models in Software Engineering; 2013 9 Oct 2013; United States.
James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning with applications in R. London: Springer; 2013.
Funding
This project was done as part of the Applied Pharmacometrics Training Fellowship, a capacity strengthening program organized and funded by Pharmacometrics Africa NPC and Certara.
Author information
Authors and Affiliations
Contributions
All authors wrote the manuscript and designed the research. I.G.A and B.S.D. performed the research and analyzed the data. All authors contributed new reagents/analytical tools.
Corresponding author
Ethics declarations
Conflict of Interest
A.L. and J.L. are employees of Eli Lilly and Company and hold stock in the company. All the other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Asiimwe, I.G., S’fiso Ndzamba, B., Mouksassi, S. et al. Machine-Learning Assisted Screening of Correlated Covariates: Application to Clinical Data of Desipramine. AAPS J 26, 63 (2024). https://doi.org/10.1208/s12248-024-00934-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-024-00934-6