Abstract
The ex vivo isolated perfused rat lung (IPL) model has been demonstrated to be a useful tool during drug development for studying pulmonary drug absorption. This study aims to investigate the potential use of IPL data to predict rat in vivo lung absorption. Absorption parameters determined from IPL data (ex vivo input parameters) in combination with intravenously determined pharmacokinetic data were used in a biopharmaceutics model to predict experimental rat in vivo plasma concentration-time profiles and lung amount after inhalation of five different inhalation compounds. The performance of simulations using ex vivo input parameters was compared with simulations using in vitro input parameters, to determine whether and to what extent predictability could be improved by using input parameters determined from the more complex ex vivo model. Simulations using ex vivo input parameters were within twofold average difference (AAFE < 2) from experimental in vivo data for all compounds except one. Furthermore, simulations using ex vivo input parameters performed significantly better than simulations using in vitro input parameters in predicting in vivo lung absorption. It could therefore be advantageous to base predictions of drug performance on IPL data rather than on in vitro data during drug development to increase mechanistic understanding of pulmonary drug absorption and to better understand how different substance properties and formulations might affect in vivo behavior of inhalation compounds.
Similar content being viewed by others
INTRODUCTION
Pulmonary drug delivery is the preferred administration route for the treatment of lung diseases such as asthma, chronic obstructive lung disease, and cystic fibrosis (1). Optimal pulmonary drug delivery of locally acting active pharmaceutical ingredients (APIs) includes high local concentration, extended lung residence time, and low systemic concentration (2). These properties enhance the pharmacological effect and decrease the dosing frequency, which improves compliance and reduces the risk of systemically adverse effects (2). To ensure the efficient and successful development of inhalation drug products, improved knowledge about the pulmonary drug absorption, i.e., dissolution, permeability, and tissue retention of the API in the lungs, is needed (3).
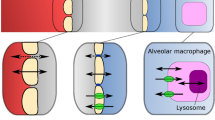
The isolated perfused rat lung (IPL) model is an ex vivo absorption model that allows parameters like epithelial permeability, solubility in epithelial lining fluid (ELF), dissolution rate, and particle wetting as well as tissue retention to be determined by computational analysis (4,5). The advantages of the IPL model over in vivo inhalation studies is that the lung-delivered dose can be closely controlled without passing the nose and the lung deposited dose can be measured directly by mass balance calculations (6,7). In addition, the effects of systemic disposition are excluded in the IPL model, because the pulmonary drug absorption is measured directly in the pulmonary vein. A physiological difference between the IPL model and the in vivo situation is that the systemic circulation, which supplies the bronchial region with blood, is not perfused in the IPL model (8). However, the drug absorption rate for solutes has previously been shown to correlate well between the IPL model and in vivo studies, which suggest that parameters obtained from the IPL model are potentially in vivo predictive (9). These advantages suggest that the IPL model may offer better opportunities than the more complex in vivo method for investigating drug absorption rate and mechanisms for solutes and different inhalation formulations.
Physiologically based biopharmaceutics (PBB) models are used to mechanistically describe absorption processes and can be used to simulate and predict drug absorption (4,5,10). These models require various drug absorption properties as input parameters, which can be either calculated from physicochemical properties or obtained from experimental measurements. For PBB models of inhaled drugs, lung absorption input parameters are commonly based on in vitro measurements (3). For example, permeability can be measured with cell monolayer models, tissue retention can be measured with tissue slices, and the dissolution rate can be based on solubility measures (11,12,13). In vitro measurements can be performed on a large scale while remaining cost-effective and are therefore well suited for predictions early in drug development (7). Some studies have shown that in vitro–based predictions can perform with high accuracy in predicting both in vivo and clinical plasma concentrations (14,15). However, there are still knowledge gaps in the use of input parameters in pulmonary absorption predictions (3). Absorption input parameters obtained from a more physiologically relevant experimental model like the IPL, might improve the predictive performance of PBB models, and also allow key factors relevant specifically to pulmonary absorption input parameters to be studied. Especially in later stages of drug development, the IPL can be useful in providing absorption data of for example different formulations.
The primary objective of this study was to investigate the potential use of IPL data to predict rat in vivo lung absorption. Absorption parameters determined from IPL data (ex vivo input parameters) were used in combination with intravenously determined pharmacokinetic data in the PBB model LungSim to predict experimental rat in vivo plasma concentration-time profiles and retained lung amount after inhalation of five different inhalation compounds. Simulations that used ex vivo input parameters were compared with simulations that used in vitro input parameters, to examine whether predictability improved when using input parameters determined from the more complex model.
METHODS
Study Drugs
Five study drugs (AZD5423, fluticasone furoate (FF), fluticasone propionate (FP), salbutamol, and salmeterol) were chosen based on their range of physicochemical properties and availability of data from the IPL model and intravenous (i.v.) and pulmonary administration of the drugs (Table I). For salbutamol and salmeterol, available inhalation data were based on studies with solutions, giving the opportunity to investigate pulmonary drug absorption without the effect of dissolution. For the low solubility APIs, AZD5423, FF, and FP, available inhalation data were from studies using suspensions; thus, the effect of dissolution could be examined.
Previously Obtained Experimental Rat IPL Data and Developed PBB Model
By applying a physiologically based biopharmaceutics (PBB) model and analyzing data obtained from the isolated perfused lung (IPL) experiments, ex vivo input parameters (permeability, solubility, and tissue retention) were obtained for the LungSim model. The following sections describe these data and methods, which were obtained and developed in previous work (4,5).
Experimental Rat IPL Data
Experimental rat IPL data were obtained from previous studies in which solutions and suspensions of the investigated APIs were administered (4,5). Briefly, the heart and lungs were isolated from the rat and put in a humidified chamber. A buffer containing albumin and glucose were used to single-pass perfuse the pulmonary circulation at a rate of 20 mL/min. The lungs were ventilated at a rate of 75 breaths per minute. The solutions (0.03 mg/mL or 0.1 mg/mL) and suspensions (1 mg/mL) were nebulized to the IPL model for 60 and 30 s, respectively. Over the course of the experiment (up to 90 min), samples were collected with an automatic sampler at predetermined time points. At the end of the experiment, the lungs were cut from the heart and trachea and were frozen for further quantitative analysis of the parent drug with the LC-MS/MS method. The lung deposited dose of API was calculated as cumulative amount in perfusate and the amount in the lungs at the end of the experiment.
IPL PBB Model
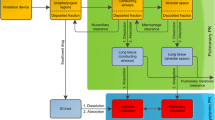
A previously developed PBB model was applied to mechanistically describe the rate and extent of pulmonary absorption based on experimental IPL data and to estimate biopharmaceutics parameters relevant to pulmonary absorption such as wetting, dissolution, permeability, and tissue retention (4,5). Briefly, the PBB model has two regionally (alveolar and tracheobronchial) specific descriptions of the deposited dose, the epithelial lining fluid (ELF), and the intracellular and vascular spaces in the lung tissue, including intra- and extracellular drug binding sites. An alveolar:tracheobronchial (Al:Tb) region lung deposition ratio of 3:2 was applied for both the solutions and suspensions (4,5). The following equations (Eqs. 1–3) were used to describe the absorption process in the lungs:
where kdiss is the rate of dissolution, k is a constant, D is the diffusion coefficient, A is the amount, Cs is the solubility in ELF, C is the concentration in ELF, h is the thickness of diffusion layer, and r is the radius of the particle. The particle radius was divided into eight bins in each region to account for the particle size distribution. The equation for the dissolution rate also takes into account the shrinking of particles, where A is raised by 2/3.
where Pmem is the membrane permeability, A and V are the amount and volume related to the donor compartment, respectively, and SAmem is the surface area of the membrane. The effective pulmonary permeability (Peff) is described as Peff = Pmem/2.
where Q is the single-pass perfusion rate (20 mL/min) and Vvasc and Avasc are the volume and amount of drug related to the vascular compartment.
The intra- and extracellular tissue retention were described with the rate constants kin and kout.
(For further information about the IPL PBB model and derivations of the above equations, see references 4 and 5.)
LungSim Model
LungSim was developed by AstraZeneca as an in-house PBB model for simulations of lung and plasma concentrations after drug administration via the pulmonary route (15,16). LungSim is an extension of GI-Sim, a previously developed biopharmaceutics tool for predictions of gastrointestinal drug absorption (17). LungSim has a lung deposition model and a lung absorption model, but because the focus of this work is on absorption input parameters, lung deposition patterns were obtained from the literature instead of estimated using the deposition model in LungSim. Absorption parameters obtained from IPL (ex vivo input parameters) or in vitro models (in vitro input parameters) were combined with systemic distribution and elimination for simulations of plasma concentrations and lung amounts (Fig. 1). Thus, the absorption input parameters (Peff, solubility and tissue retention) differed between the two settings (ex vivo and in vitro) while formulation-specific parameters (particle size and deposition pattern) and pharmacokinetic parameters (plasma clearance, the volume of distribution, fraction unbound in plasma and distribution parameters) applied in the LungSim model were the same for both settings (Table II).
Ex Vivo Input Parameters
The IPL PBB model is similar to the LungSim PBB model, but has a few key differences:
-
The applied volumes and areas of the alveolar and tracheobronchial region are different between the two models.
-
The Peff value is scaled based on the thickness of the epithelium in the LungSim model, resulting in regional values for Peff. In the IPL PBB model, the Peff value is not scaled (i.e., the same value was applied in the alveolar and tracheobronchial region).
-
The vascular space is not divided into alveolar and a tracheobronchial region as it is in the IPL PBB model (4).
-
The dissolution rate is slightly differently described in LungSim than the IPL PBB model.
-
Removal of drug particles from ELF by mucociliary clearance exists in LungSim but not in the IPL PBB model.
These differences needed to be assessed to ensure that the ex vivo input parameters represented the absorption in the experimental IPL model when used in the LungSim model.
To correct for these differences, Peff and tissue retention (kin and kout) were re-estimated using the previously developed IPL PBB model but applying the same volumes and surface area as in LungSim (Phoenix® WinNonlin® 8.1, Certara USA, NJ, USA) (4). The difference in the division of vascular space was not corrected for because it did not affect the absorption rate (data not shown). (To account for parameter specific differences, see the sections below.)
Permeability
The re-estimated pulmonary Peff was used as an input value for lung Peff (4) (Table II). In opposition to default settings, no scaling of the Peff value between alveolar and bronchial regions was adopted in the LungSim model, by analogy with the modeling strategy for estimation of this parameter.
Tissue Retention
The re-estimated values for kin and kout were used as input values for tissue retention in LungSim (4) (Table II). These values describe tissue retention as a dynamic process, where kin is the rate of distribution into the tissue and kout is the rate of distribution out of the tissue. The LungSim model requires a fu,tissue factor, calculated as:
Solubility
As mentioned, the dissolution is described slightly differently in the IPL PBB model and the LungSim model. LungSim allows estimation of solubility based on a dissolution concentration-time profile, which was performed to ensure a correct ex vivo input value for solubility. Dissolution profiles from the IPL experiments were obtained by applying the IPL PBB model.
As previously reported, the absorption rate in the IPL model for the suspension and solution of AZD5423 was the same, indicating that dissolution was not rate-limiting for the suspension (with a similar particle size distribution as used in the in vivo validation data) and a dissolution profile not possible to obtain (5). Therefore, AZD5423 was in this study simulated as a solution in LungSim even if administered as a suspension.
In Vitro Input Parameters
Permeability
Intestinal Peff was used as the in vitro input parameter for pulmonary Peff (Table II). The intestinal Peff was calculated from a Caco-2 apparent permeability (Papp) − intestinal Peff correlation (intestinal Peff = 1.8 Papp + 1.06, human UWL thickness = 8.6 × 10−5, Caco-2 UWL thickness 7.5 × 10−4 (17,18)). The Caco-2 measurements were performed with major drug influx and efflux transporters inhibited. Values for Caco-2 Papp were obtained from Eriksson et al. (4). The scaling of pulmonary Peff in LungSim was kept as default for the simulations using in vitro input parameters.
Tissue Retention
The drug fraction unbound in the lung (fu,tissue) was used as the parameter for describing tissue retention and was obtained using volume unbound (Vu,lung) measured in lung slices as described by Bäckström et al. (11), where fu,tissue = 1/Vu,lung× Vu,lung values were obtained from AstraZeneca in-house database (Table II). The tissue distribution settings in LungSim were set to “equilibrium” to accommodate for this input parameter, and so distribution time dependencies were not evaluated in these simulations.
Solubility
The input value for in vitro solubility was the solubility in phosphate buffer at pH 7.4, as reported by Eriksson et al. (5) (Table II).
Pharmacokinetic Input Parameters
Intravenous plasma concentration-time profiles for all APIs were obtained from AstraZeneca in-house database. The in-house studies were approved by the local ethics committee. The i.v. bolus doses administered to the rats were 0.924 and 0.802 μmol/kg for AZD5423, 0.943 and 0.922 μmol/kg for FF, 1.03 and 0.949 μmol/kg for FP, 4.48 μmol/kg for salbutamol (three replicates) and 2.43 μmol/kg for salmeterol (two replicates). Plasma samples were retrieved at predetermined time points over 24 h for AZD5423, FF, and FP, over 6 h for salbutamol and over 12 h for salmeterol. The i.v. profiles were analyzed by applying a two-compartment model to obtain pharmacokinetic parameters describing the systemic distribution and elimination, i.e., volume of distribution (Vd), rate of distribution (k12 and k21), and plasma clearance (CL) (Phoenix® WinNonLin® 8.1, Certara USA, NJ, USA) (Table II). Estimated pharmacokinetic parameters were used to describe systemic distribution and elimination in the LungSim model.
Description of In Vivo Validation Dataset
The in vivo validation dataset was obtained from AstraZeneca in-house database and included both nose-only inhalation and intratracheal instillation data. The in-house studies were approved by the local ethics committee.
In Vivo Nose-Only Inhalation Data
AZD5423, FF, and FP were administered as nose-only inhalation to rats. The lung-delivered dose was calculated to be between 23 and 25 μg/kg for the APIs. The APIs were nebulized as suspensions over 30 min. Measurements of droplet and particle size distributions were performed in-house at AstraZeneca (Table II). The median particle size and span of the particles were adjusted to represent the lung-delivered particle size distribution by only considering the particles ≤ 1 μm to deposit in the lungs (Table II). This assumption was based on the study by Schmid et al. (19), where particles ≥ 1 μm to a high extent deposit in the nose and extrathoracic region. The regional deposition (Al:Tb ratio) for nose-only inhalation was assumed to be 50:50 (20). Both plasma concentration and lung amount were available for the inhalation data.
In Vivo Intratracheal Instillation Data
Lung-delivered doses of 20 μg/kg of salbutamol and salmeterol were administered to rats as an intratracheal instillation (i.t.) (LDD in Table II). The APIs were administered as solutions. The Al:Tb ratio for i.t. administration was assumed to be 30:70 (21). Only plasma concentration was available for the i.t. data.
Simulations
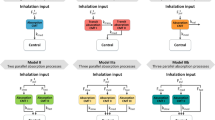
Simulations for each API were performed in LungSim with the ex vivo and in vitro input parameters summarized in Table II. For each API, additional simulations were performed using ex vivo input parameters in which the bronchial permeability values were doubled. This was done to compensate for the possibility of lower bronchial absorption in the IPL model than in vivo, because the tracheobronchial region is not perfused in the IPL model. Because deposition pattern is an uncertain parameter in predictions of lung absorption, higher Al:Tb ratios were also applied (80:20 for AZD5423, FF, and FP and 50:50 for salbutamol and salmeterol) to test the effect.
Statistical Evaluation
The similarity between simulated and experimental data was evaluated by comparing the area under the curve (AUC), maximum concentration (Cmax), time when Cmax occurred (tmax), absolute average fold error (AAFE), and average fold error (AFE). AUC, Cmax, and tmax were calculated using GraphPad Prism 8® (GraphPad Software, San Diego, USA). AAFE and AFE were defined as follows:
where sim is the simulated data, expe is the experimental data, and N is the number of data points included. An AAFE value of 1 indicates a perfect agreement between simulated and experimental data, while a value of 2 indicates an average twofold difference between the compared data. AAFE can therefore be used to evaluate the difference between two datasets and AFE evaluates whether the simulation over- or underestimates the experimental data.
RESULTS
Simulations Using Ex Vivo Input Parameters
The simulations using ex vivo input parameters were within twofold average difference (AAFE < 2) from the experimental in vivo data for all APIs except FP (Fig. 2, Table III). In general, the simulations had a lower Cmax than the experimental in vivo data (Table III). The simulated lung amount was higher than the experimental lung amount for all APIs (Fig. 3). The simulations with higher bronchial permeabilities and a higher Al:Tb ratio gave a comparatively similar plasma concentration-time profile (difference in tmax < 0.1 h and Cmax < 20%) (Fig. 2, Table III). With these modified simulation settings, the predicted Cmax improved for AZD5423, FP, and salmeterol, and AAFE improved for AZD5423, FP, and salbutamol compared with the initial simulation settings (Table III). In addition, the modified simulation settings improved the predictions of lung amount for all APIs (Fig. 3). Absorption in the alveolar region was in general rapid (> 90% of drug absorbed within 1 h), while it was slower in the tracheobronchial region (Fig. 4). AZD5423 permeated the epithelial membrane and was then retained in the tissue (the slope of the amount in tissue over time is less steep than the slope of the amount in solution over time, Fig. 4). FF and FP dissolved slowly in the tracheobronchial region; FF was retained in the tissue after permeation while FP was not (Fig. 4). For salbutamol, permeation was the rate-limiting step, while for salmeterol, tissue retention was rate-limiting (Fig. 4).
Simulations Using In Vitro Input Parameters
The simulations using in vitro absorption parameters had AAFEs > 2 compared with experimental in vivo data for all APIs except salmeterol (Table III). In general, the simulations under predicted the rate of pulmonary absorption (Figs. 5 and 6). The in vitro simulations were less accurate in predicting Cmax than the simulations using ex vivo input parameters for all APIs (Fig. 5) and had a higher AAFE for all APIs except salmeterol (where the two AAFEs were nearly equal, Table III). For salmeterol, the ex vivo input parameters predicted the Cmax better, while the in vitro input parameters gave a better prediction for the later time points of plasma concentration (Fig. 5). The dissolution rate and tissue retention in the tracheobronchial region were the rate-limiting steps for the absorption of AZD5423, FF, and FP (Fig. 4). For salbutamol and salmeterol, tissue retention in the tracheobronchial region was rate-limiting (Fig. 4).
DISCUSSION
Lung-specific absorption parameters obtained via the IPL model were used to simulate rat in vivo plasma concentration-time profile and lung amount for five different inhalation compounds. Simulations were within 2-fold absolute average error (AAFE) of the experimental in vivo rat data, with the exception of fluticasone propionate, indicating that drug absorption parameters obtained from the IPL model are predictive of in vivo lung absorption. Simulations using in vitro input parameters were compared with simulations using ex vivo input parameters, and those based on ex vivo parameters were significantly more accurate (as indicated by lower AAFE values) for the investigated APIs (except for salmeterol where the difference in AAFE was insignificant). These results demonstrate the advantage of the IPL method over in vitro methods for determining input parameters for predictions of in vivo plasma concentration-time profile and lung amount. In contrast to in vitro models, the IPL model resembles the in vivo dissolution better because it offers physiologically relevant volume, fluid composition, and sink/non-sink conditions. IPL also resembles in vivo permeability better because the epithelial membrane is the same as would be found in an in vivo experiment; for similar reasons, IPL resembles in vivo tissue retention better than in vitro because it offers the same volume, tissue composition, and dynamic binding processes. In addition, IPL includes the diversity and complexity of lung structure, including differences in the abovementioned parameters between the alveolar and tracheobronchial regions.
IPL is an advanced ex vivo model closely mimicking the in vivo lung physiology because it includes continuous lung breaths and the lung is perfused. One significant difference between IPL and in vivo experiments is that the bronchial region is not perfused in IPL; however, absorption parameters obtained from IPL have been shown to be predictive of in vivo lung absorption. Because the amount absorbed in IPL has reached near-complete absorption in several studies, it has been suggested that drug can be absorbed from the bronchial region, despite this region is not being perfused (4,8). Such a result might be explained by drug transport from the bronchial region through anastomoses and/or diffusion to the alveolar capillaries (22). Increasing permeability in the tracheobronchial region for the simulations improved predictions (lower AAFE) for three out of five investigated APIs. Based on that result, it seems that the lack of bronchial perfusion in IPL does not necessarily slow absorption in this region, lending further evidence that IPL successfully mimics the in vivo condition.
The absorptive surface area in the tracheobronchial region is much smaller than in the alveolar region, which is why simulated absorption from the tracheobronchial region was significantly slower than the simulated absorption from the alveolar region for the studied drugs (Table II). The difference in absorption between the two lung regions was even greater for the simulations using in vitro input parameters, because the pulmonary Peff in that case was scaled with the thickness of the epithelium (Table II).
The in vitro Peff value was calculated from a correlation between human intestinal Peff and Caco-2 Papp, which has often been used for biopharmaceutics intestinal absorption prediction, while the ex vivo Peff value was estimated from IPL data using the IPL PBB model (23). The two methods used to obtain a Peff value rendered significantly different values, especially when comparing the values for the alveolar region. The effect of this difference in Peff value between simulations using in vitro or ex vivo input parameter could be seen for the alveolar absorption of solutions, which was much higher for the in vitro simulations (Fig. 4). However, the high in vitro Peff value was compensated by a high tissue retention for salmeterol, resulting in a slower overall absorption rate. Similarly, Peff would have a low impact on the overall absorption rate if dissolution is the rate-limiting step, which is the case for low solubility APIs. The difference in Peff was smaller in the tracheobronchial region because of the scaling of the value in the in vitro simulations. Scaling of Peff value between the lung regions was attempted in the IPL PBB model but this revised model was not able to explain the experimental data obtained from IPL (data not shown). In the simulations using in vitro input parameters, both salbutamol and salmeterol were absorption limited by retention in the lung tissue in the tracheobronchial region despite a big difference in fu,tissue between the two APIs. This indicates that the permeability on the basolateral side of the membrane (i.e., transport from the membrane into the bloodstream) could be rate-limiting for APIs with low permeability and high fu,tissue. The effect of the permeability value on the overall absorption rate of solutions can be seen when comparing the ex vivo simulation of AZD5423 to the simulations of salbutamol and salmeterol. AZD5423, with a higher Peff value, has a higher absorption rate in both lung regions compared with salbutamol and salmeterol.
The setting for tissue retention differed between simulations using ex vivo and in vitro input parameters because of the nature of the experimental data. Dynamic distribution was applied for the ex vivo simulations and equilibrium distribution was applied for in vitro simulations. When lung tissue distribution is fast, as for example with AZD5423, no significant difference between the two settings was observed. In contrast, for APIs like salbutamol with slow distribution, significant differences between the settings will occur (data not shown). For example, if an equilibrium distribution is applied, salbutamol will be highly retained in the tissue because fu,tissue is very low, while if a dynamic distribution is applied, retention will be low because the rate of distribution is low (Table II). Predictiveness may be improved by measuring dynamic distribution in lung slices rather than equilibrium distribution, which will also better represent the actual absorption process (11,24,25).
The solubility used in the in vitro simulations was lower than in the ex vivo simulations, which resulted in a higher amount of API remaining as solid in the in vitro simulations (Fig. 4, Table III). In the in vitro simulations of APIs administered as suspensions (where dissolution was the rate-limiting step), the applied solubility seemed to be too low, as indicated by the low values for Cmax and AUC. Solubility in phosphate buffer pH 7.4 might not represent solubility in lung lining fluid, and a more optimal input value for solubility needs to be found if simulations and models are to be improved.
The systematic exposure predictions in Fig. 2 based on the ex vivo input parameters performed well in predicting in vivo lung absorption, although they have ex vivo model origin. However, predicted and observed values were not identical, and additional investigation is required. One possible source of deviation is that the IPL and in vivo models used different administration techniques and inhalation devices, which would result in different deposition patterns in the lungs, which in turn would likely affect absorption (26). The deposition pattern was an approximate estimation for both the in vivo and IPL data, and so simulations using different deposition patterns were also performed to test the effect on simulated absorption. Deposition pattern did affect the plasma concentration, especially Cmax, and clearly, it is important to have a good estimate of deposition pattern; however, there is little experimental data to validate any given pattern (27).
Another possible source of difference is that the API batches were not identical for IPL and in vivo experiments, which could affect the dissolution rates. Differences in particle size distribution were accounted for in the simulations, but differences in other factors like shape and agglomeration behavior could not be accounted for because there was no available data on these factors. Another potentially important difference is that the lungs in IPL are excised from the rat body and are therefore composed of dying tissue. This tissue has been shown to be viable for several hours, but the loss in viability over time might alter absorption compared with the living tissue in vivo (28). Finally, it is difficult to simulate and predict in vivo plasma concentration-time profiles when the in vivo data are obtained from nose-only inhalation experiments, because the lung-delivered dose can only be an approximate estimate. Both the simulations using ex vivo and in vitro input parameters had lower Cmax and AUC values than the in vivo data for two out of three of the APIs administered with nose-only inhalation, which suggests that the dosage applied in the simulations might be too low.
CONCLUSIONS
This study has further demonstrated the usefulness of data obtained with IPL by showing that absorption parameters obtained by this method yield better predictions of rat in vivo lung absorption of both solution and suspension formulations than absorption parameters determined from standard in vitro measurements. It would be advantageous to use predictions based on IPL data during drug development in order to increase mechanistic understanding of the pulmonary drug absorption processes and to better predict how changes in drug substance properties and formulation will affect the in vivo performance of inhalation compounds.
Change history
08 December 2020
A Correction to sthis paper has been published: https://doi.org/10.1208/s12248-020-00545-x
References
Labiris N, Dolovich M. Pulmonary drug delivery. Part I: physiological factors affecting therapeutic effectiveness of aerosolized medications. Br J Clin Pharmacol. 2003;56(6):588–99.
Patton JS, Byron PR. Inhaling medicines: delivering drugs to the body through the lungs. Nat Rev Drug Discov. 2007;6(1):67–74.
Bäckman P, Arora S, Couet W, Forbes B, de Kruijf W, Paudel A. Advances in experimental and mechanistic computational models to understand pulmonary exposure to inhaled drugs. Eur J Pharm Sci. 2018;113:41–52.
Eriksson J, Sjögren E, Thörn H, Rubin K, Bäckman P, Lennernäs H. Pulmonary absorption–estimation of effective pulmonary permeability and tissue retention of ten drugs using an ex vivo rat model and computational analysis. Eur J Pharm Biopharm. 2018;124:1–12.
Eriksson J, Thorn H, Sjögren E, Holmstén L, Rubin K, Lennernäs H. Pulmonary dissolution of poorly soluble compounds studied in an ex vivo rat lung model. Mol Pharm. 2019;16(7):3053–64.
Mehendale HM, Angevine LS, Ohmiya Y. The isolated perfused lung—a critical evaluation. Toxicology. 1981;21(1):1–36.
Sakagami M. In vivo, in vitro and ex vivo models to assess pulmonary absorption and disposition of inhaled therapeutics for systemic delivery. Adv Drug Deliv Rev. 2006;58(9–10):1030–60.
Tronde A, Nordén B, Jeppsson A-B, Brunmark P, Nilsson E, Lennernäs H, et al. Drug absorption from the isolated perfused rat lung-correlations with drug physicochemical properties and epithelial permeability. J Drug Target. 2003;11(1):61–74.
Tronde A, Nordén B, Marchner H, Wendel AK, Lennernäs H, Bengtsson UH. Pulmonary absorption rate and bioavailability of drugs in vivo in rats: structure–absorption relationships and physicochemical profiling of inhaled drugs. J Pharm Sci. 2003;92(6):1216–33.
Sjögren E, Tammela TL, Lennernäs B, Taari K, Isotalo T, Malmsten L-Å, et al. Pharmacokinetics of an injectable modified-release 2-hydroxyflutamide formulation in the human prostate gland using a semiphysiologically based biopharmaceutical model. Mol Pharm. 2014;11(9):3097–111.
Bäckström E, Lundqvist A, Boger E, Svanberg P, Ewing P, Hammarlund-Udenaes M, et al. Development of a novel lung slice methodology for profiling of inhaled compounds. J Pharm Sci. 2016;105(2);838-84.
Riley T, Christopher D, Arp J, Casazza A, Colombani A, Cooper A, et al. Challenges with developing in vitro dissolution tests for orally inhaled products (OIPs). AAPS PharmSciTech. 2012;13(3):978–89.
Manford F, Tronde A, Jeppsson A-B, Patel N, Johansson F, Forbes B. Drug permeability in 16HBE14o-airway cell layers correlates with absorption from the isolated perfused rat lung. Eur J Pharm Sci. 2005;26(5):414–20.
Bäckman P, Tehler U, Olsson B. Predicting exposure after oral inhalation of the selective glucocorticoid receptor modulator, AZD5423, based on dose, deposition pattern, and mechanistic modeling of pulmonary disposition. Journal of aerosol medicine and pulmonary drug delivery. 2017;30(2):108–17.
Tehler U, Fransson R, Thorn H, Franek F, Westergren J. Lung-Sim: a physiologically based biopharmaceutical prediction tool. Journal of Aerosol Medicine and Pulmonary Drug Delivery 2018: Mary Ann Liebert, INC 140 Huguenot Street, 3RD FL, New Rochelle, NY 10801 USA.
Thorn H, Loof T, Petersson G, Franek F, Fransson R, Tehler U. Deposition modelling of inhaled aclidinium and budesonide and comparison to gamma scintigraphy measurement. Journal of Aerosol Medicine and Pulmonary Drug Delivery 2018: Mary Ann Liebert, INC 140 Huguenot Street, 3RD FL, New Rochelle, NY 10801 USA.
Sjögren E, Westergren J, Grant I, Hanisch G, Lindfors L, Lennernäs H, et al. In silico predictions of gastrointestinal drug absorption in pharmaceutical product development: application of the mechanistic absorption model GI-Sim. Eur J Pharm Sci. 2013;49(4):679–98.
Fredlund L, Winiwarter S, Hilgendorf C. In vitro intrinsic permeability: a transporter-independent measure of Caco-2 cell permeability in drug design and development. Mol Pharm. 2017;14(5):1601–9.
Schmid O, Bolle I, Harder V, Karg E, Takenaka S, Schulz H, et al. Model for the deposition of aerosol particles in the respiratory tract of the rat. I. Nonhygroscopic particle deposition. Journal of aerosol medicine and pulmonary drug delivery. 2008;21:291–308.
Raabe OG, Al-Bayati MA, Teague SV, Rasolt A. Regional deposition of inhaled monodisperse coarse and fine aerosol particles in small laboratory animals. Inhaled Particles VI: Elsevier; 1988: 53–63.
Codrons V, Vanderbist F, Ucakar B, Préat V, Vanbever R. Impact of formulation and methods of pulmonary delivery on absorption of parathyroid hormone (1–34) from rat lungs. J Pharm Sci. 2004;93(5):1241–52.
Deffebach ME, Charan NB, Lakshminarayan S, Butler J. The bronchial circulation: small, but a vital attribute of the lung 1–3. Am Rev Respir Dis. 1987;135(2):463–81.
Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology. 1989;96(2):736–49.
Macintyre AC, Cutler DJ. The potential role of lysosomes in tissue distribution of weak bases. Biopharm Drug Dispos. 1988;9(6):513–26.
Miller-Larsson A, Jansson P, Runstrom A, Brattsand R. Prolonged airway activity and improved selectivity of budesonide possibly due to esterification. Am J Respir Crit Care Med. 2000;162(4):1455–61.
Salar-Behzadi S, Wu S, Mercuri A, Meindl C, Stranzinger S, Fröhlich E. Effect of the pulmonary deposition and in vitro permeability on the prediction of plasma levels of inhaled budesonide formulation. Int J Pharm. 2017;532(1):337–44.
Lizal F, Jedelsky J, Morgan K, Bauer K, Llop J, Cossio U, et al. Experimental methods for flow and aerosol measurements in human airways and their replicas. Eur J Pharm Sci. 2018;113:95–131.
Niemeier RW, Bingham E. An isolated perfused lung preparation for metabolic studies. Life Sci. 1972;11(16):807–20.
Irwin JJ, Shoichet BK. ZINC− a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–82.
Acknowledgments
This study is a part of the science program of the Swedish Drug Delivery Forum (SDDF).
Funding
Animal Ethics Committee of Gothenburg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The in-house studies were approved by the Animal Ethics Committee of Gothenburg.
Conflict of Interest
The other authors have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: There was a conversion error when processing the PDF version of this article. The online version is correct, however the equations in the PDF, all “=” symbols were inadvertently changed to “0”.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eriksson, J., Sjögren, E., Lennernäs, H. et al. Drug Absorption Parameters Obtained Using the Isolated Perfused Rat Lung Model Are Predictive of Rat In Vivo Lung Absorption. AAPS J 22, 71 (2020). https://doi.org/10.1208/s12248-020-00456-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-020-00456-x