Abstract
Background
Despite the known role of mitosis in colorectal cancer, previous associations of long-term aspirin use with suppressed cancer-related epigenetic aging did not involve epigenetic mitotic clocks. We investigated these relationships using three epigenetic mitotic clocks developed for cancer risk prediction: EpiTOC, EpiTOC2, and MiAge. We utilized publicly available HumanMethylationEPIC BeadChip data from 112 healthy colon (proximal and distal) mucosal samples taken at baseline (T1) and at 10-years follow-up (T2) from a screening cohort of 28 Polish women (11 non-users and 17 long-term [≥ 2 years] aspirin users). Mitotic clock values were divided by chronological age at each timepoint to obtain intrinsic rates (IRs). We evaluated differences in residuals of the mitotic clock IRs taken from linear mixed effects models adjusted for BMI, polyp status, and DNA methylation batch.
Findings
EpiTOC, EpiTOC2, and MiAge were significantly correlated with chronological age (P < 0.05) with correlations ranging from 0.41 to 0.63. The EpiTOC, EpiTOC2, and MiAge clocks were strongly correlated with each other in proximal and distal samples (r > 0.79, P < 0.0001). We observed proximal within group median clock IR deceleration for EpiTOC (-0.0004 DNAm, P = 0.008), EpiTOC2 (− 16 cell divisions, P = 0.009), and MiAge (− 3 cell divisions, P = 0.002) for long-term aspirin users from T1 to T2 but not for non-users. In distal samples, only the long-term user MiAge IR was significantly deaccelerated (− 3 cell divisions, P = 0.009).
Conclusions
Our observed findings support previously reported longitudinal associations of aspirin use with deceleration of other epigenetic age measures in the proximal colon. Our mitotic clock results suggest that cell proliferation could play a role in some aspirin relationships with epigenetic aging. Furthermore, the findings provide added impetus for establishing gold standards for epigenetic aging and consensus guidelines for more comprehensive reporting in future epigenetic aging cancer studies.
Similar content being viewed by others
Introduction
Existing data, including evidence from human randomized controlled trials, suggests that aspirin may be protective against colorectal cancer (CRC) in certain populations [1,2,3]. In accordance with this evidence, the United States Preventative Services Task Force (USPSTF) has issued guidelines recommending the use of low-dose aspirin for the primary prevention of CRC in adults aged 50–59 years with a ≥ 10% 10-year cardiovascular disease risk [4]. Despite the optimism surrounding the role that aspirin may play as a CRC chemopreventive agent, issues of long-term patient drug adherence, therapeutic contraindications, and CRC’s existing immense morbidity—the third most common cancer diagnosed globally with an estimated 1.8 million new cases annually [5]—guarantee a continued need for disease screening and surveillance [1]. For this reason, it will be important to characterize the relationship of aspirin use with CRC screening and surveillance modalities.
Measures of epigenetic aging are among the potential, novel modalities for CRC screening and surveillance [6, 7]. These DNA methylation-based metrics, sometimes called “epigenetic clocks,” are believed to represent biological, rather than chronological, aging and can offer better insights regarding an individual’s present and future morbidity and mortality risks [8, 9]. To date, blood epigenetic age acceleration (Horvath’s pan-tissue metric [10]) has been associated with future colon cancer development [11], and survival associated epigenetic age acceleration (Horvath metric) has also been demonstrated in colon tumors [12]. Moreover, a more recent 10-year study of healthy colon tissue in Polish women reported epigenetic age deceleration (PhenoAge metric [13]) in the proximal colon of long-term aspirin users when compared to non-users [14]. Although these studies do take advantage of the most well-known epigenetic age measures, they do not include epigenetic aging measures that estimate cell divisions—otherwise known as “mitotic clocks.” Understanding aspirin and CRC relationships with mitotic clocks is particularly important given that deviations from chronological age captured by many of the other epigenetic clocks are independent of cell proliferation [14]. This may result in the conclusion that aspirin’s relationship with epigenetic aging processes is solely proliferation independent when this may not be the case. Studying these relationships may also offer insights for furthering the mechanistic understanding of aspirin’s chemopreventive potential. The known role of mitosis in CRC provides additional impetus for this mitotic clock analysis [15].
We identified three mitotic clock measures in the literature: Epigenetic Time to Cancer (EpiTOC), Epigenetic Time to Cancer-2 (EpiTOC2), and Mitotic Age (MiAge) [16,17,18]. EpiTOC is the average DNA methylation over its 385 component CpG sites and represents the age-cumulative increase in DNA methylation due to putative cell-replication errors at these sites [16]. Furthermore, EpiTOC leverages CpG sites localized to Polycomb group target genes, which are generally unmethylated in many fetal tissues but gain methylation during aging and ontogeny. The value it provides is a relative estimate of the number of stem cell divisions per stem cell. The EpiTOC2 biomarker is more robust than EpiTOC because it estimates the cumulative number of stem cell divisions in a tissue—yielding direct mitotic count estimates [17]. Increases in EpiTOC and EpiTOC2 have been demonstrated in various cancers, pre-invasive cancer lesions (including colorectal adenomas), and normal buccal tissues exposed to cigarette smoke (a known carcinogen) [16, 17]. MiAge is designed to represent the total number of lifetime cell divisions of a tissue and was found to be universally accelerated in 13 cancer types (including colon adenocarcinoma) when tumors were compared to adjacent normal tissues [18]. Accelerated MiAge was also found to be associated with worse cancer survival [18].
In the present analysis, we utilize the aforementioned Polish healthy colon dataset [14] and explored relationships of long-term aspirin use with EpiTOC, EpiTOC2, and MiAge. Additionally, we take an inventory of existing CRC epigenetic aging studies and provide recommendations for future work in this area.
Methods
Study population and data collection
We conducted analyses using a publicly available NCBI GEO MethylationEPIC BeadChip healthy colon mucosa dataset (Series GSE142257, updated on “https://www.ncbi.nlm.nih.gov/geo” on November 9, 2020). The dataset is comprised of 124 normal cecum (proximal) and sigmoid (distal) colon mucosa biopsies from 31 healthy Polish women and was collected for a previous longitudinal analysis of aspirin use and epigenetic aging [14]. Colonoscopies with mucosa biopsies were performed on the women at baseline (T1) and during follow-up 10 years later (T2). In total, each study participant had four biopsies, proximal and distal biopsies at T1 and proximal and distal biopsies at T2. Aspirin use at baseline (T1) and follow-up (T2) was assessed via a self-administered questionnaire and was classified as non-user or long-term user (≥ twice per week for ≥ 2 years). Of the 31 study participants, 17 were long-term users at T1 and 20 were long-term users at T2—three non-users from T1 switched to long-term users at T2. Given that this study aimed to examine the longitudinal relationship of aspirin use with epigenetic mitotic clocks over the ten year study period, those three individuals with cross-over aspirin use were excluded from the analyses leaving 28 subjects (112 mucosa samples). The dataset also contained information on participant chronological ages, BMIs, DNA methylation batch, and if any type of polyps were observed on T1 or T2 colonoscopies (yes versus no). Additional details about study participants as well as informed consent and ethical conduct of study information regarding the collection of this data can be found in the original publication of the data [14].
DNA methylation data and epigenetic mitotic clocks
Per the initial publication of this dataset, high-definition colonoscopy with narrow-band imaging was used to obtain normal mucosa biopsies and genomic DNA was extracted from these fresh biopsies. Genome-wide DNA methylation profiling was then performed using the Illumina Infinium MethylationEPIC BeadChip [14]. Minfi preprocessed and normalized DNA methylation data was downloaded from NCBI by the authors of the present manuscript. From a literature review, we identified three robust epigenetic mitotic clocks designed to use MethylationEPIC BeadChip data: EpiTOC [16], EpiTOC2 [17], and MiAge [18]. EpiTOC and EpiTOC2 were calculated from the downloaded methylation beta values using instructions and R code available at https://doi.org/10.5281/zenodo.2632938. Similarly, MiAge was also calculated from the downloaded methylation beta values using instructions and R code available at http://www.columbia.edu/~sw2206/softwares.htm.
Statistical analysis
We conducted our primary analyses using age-independent intrinsic rates (IRs) for each mitotic clock. As described in the literature [17], these IRs were calculated by dividing the mitotic clock values for each participant by their respective chronological age. To then examine if mitotic clock IRs varied between non-users and long-term users between our two timepoints, independent of other covariates, we created acceleration residuals for each measure. More specifically, IR residuals were obtained by regressing IRs for each mitotic clock from all 112 samples on baseline and follow-up BMI, polyp status, and DNA methylation batch in linear mixed effects models. The models also included a random intercept for participants to account for repeated measures from each participant. We then conducted within group, paired Wilcoxon signed rank tests of EpiTOC, EpiTOC2, and MiAge IR residuals between T1 and T2 timepoints for non-users and long-term aspirin users respectively. Lastly, we performed analyses to determine if there were between group, statistically significant differences in mitotic clock IR changes from T1 to T2 (comparing non-users to long-term aspirin users). For non-users and long-term aspirin users respectively we subtracted mitotic clock values at T1 from T2. We then performed Wilcoxon rank sum tests to compare these “difference” values between the two groups of aspirin use. All analyses were repeated for EpiTOC, EpiTOC2, and MiAge residuals (not divided by chronological age). All statistical analyses were performed using R Version 3.6.3 (R Core Team, Vienna, Austria), and a P value < 0.05 was used as the threshold for statistical significance.
Results
Table 1 presents the baseline (T1) and follow-up (T2) clinical and mitotic clock characteristics of the study participants. All participants were women and 39% (11) were non-users of aspirin throughout the 10-year duration of the study. At baseline and follow-up, non-users had a mean (SD) age of 53.8 (6.7) and 63.8 (6.7) years respectively. At baseline and follow-up, long-term aspirin users had a mean (SD) age of 60.2 (5.7) and 70.2 (5.7) years respectively. Baseline median chronological ages for non-users and long-term aspirin users were 50 and 60 years respectively (Wilcoxon rank sum test P = 0.01). At baseline, the majority of non-users had polyps observed on colonoscopy (64%) and had a high BMI (≥ 26 kg/m2) (55%). At follow-up, the majority of non-users did not have polyps on colonoscopy (91%). At baseline, the majority of long-term aspirin users had a normal BMI (between 8.5 to 25 kg/m2) (59%) and did not have polyps on colonoscopy (76%). Similar patterns were observed for long-term users at follow-up.
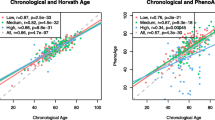
All median EpiTOC, EpiTOC2, and MiAge values for non-users were lower when compared to values for long-term users at each timepoint, not accounting for baseline age. In non-users from T1 to T2, proximal and distal colon EpiTOC, EpiTOC2, and MiAge median values increased. In long-term aspirin users from T1 to T2, proximal MiAge median values decreased. All other median values for long-term aspirin users increased from T1 to T2. Figure 1 presents the correlations of EpiTOC, EpiTOC2, and MiAge with chronological age for proximal and distal samples at each timepoint in non-users and long-term users. All correlations were statistically significant (P < 0.05) with correlation coefficients ranging from 0.41 to 0.63. The strongest correlations with chronological age were observed with EpiTOC and MiAge in the distal colon samples. All correlations between EpiTOC, EpiTOC2, and MiAge were strong in proximal (r > 0.89, P < 0.0001) and distal (r > 0.79, P < 0.0001) samples at T1 and T2. All correlations between EpiTOC and EpiTOC2 were particularly strong in both colon locations and at both time points (r > 0.99, P < 0.0001).
Table 2 presents the median mitotic clock IR residuals by timepoint, aspirin use, and colon location. On comparison, median T2 IR residuals were lower than median T1 IR residuals for both colon locations and in both non-users and long-term aspirin users. Nevertheless, this deceleration was statistically significant in the proximal colon of long-term aspirin users. We observed median clock IR deceleration for EpiTOC (− 0.0004 DNAm, P = 0.008), EpiTOC2 (− 16 cell divisions, P = 0.009), and MiAge (− 3 cell divisions, P = 0.002) in long-term aspirin users from T1 to T2. We also observed median clock IR deceleration for distal colon MiAge (− 3 cell divisions, P = 0.009) in long-term aspirin users from T1 to T2. Figure 2 graphically depicts these results using boxplots of the mitotic clock IR residuals by timepoint and long-term aspirin use. In results of EpiTOC (0.013 DNAm, P = 0.002), EpiTOC2 (796 cell divisions, P = 0.003), and MiAge (207 cell divisions, P = 0.007) residuals not accounting for chronological age, the opposite trend of mitotic clock acceleration in distal long-term user colon was observed (Table S1). There was also a general trend of higher median T2 residuals when compared to median T1 residuals in these results.
Mitotic clock intrinsic rate distribution relationships. Boxplots of mitotic clock age-adjusted intrinsic rate (IR) residuals (from models additionally adjusted for BMI, polyp status, and methylation batch) by aspirin use and colon location for the 10-year follow-up period for EpiTOC (A), EpiTOC2 (B), and MiAge (C). P values are from paired Wilcoxon signed rank tests comparing T2 and T1 for each aspirin use group. T1, timepoint 1 (baseline). T2, timepoint 2 (10-year follow-up)
Table 3 presents the baseline versus 10-year follow-up comparisons of median mitotic clock IR residual differences between non-users and long-term aspirin users. For all clocks and in proximal colon, long-term users had a more negative median difference in T2 minus T1 IR residuals when compared to non-users. Nonetheless, none of these relationships meet the threshold for statistical significance. Figure 3 graphically depicts these results using boxplots. Table S2 presents the results for the same analysis using EpiTOC, EpiTOC2, and MiAge residuals not accounting for chronological age. Here, the results do not meet statistical significance and trends are less discernable.
Aspirin use mitotic clock intrinsic rate difference distributions. Boxplots of the differences (T2 − T1) in mitotic clock age-adjusted intrinsic rate (IR) residuals (from models additionally adjusted for BMI, polyp status, and methylation batch) by aspirin use and colon location for EpiTOC (A), EpiTOC2 (B), and MiAge (C). P values are from Wilcoxon rank sum tests comparing differences (T2 − T1) between non-users and long-term aspirin users. T1, timepoint 1 (baseline). T2, timepoint 2 (10-year follow-up)
Discussion
In this analysis of healthy colon DNA methylation data spanning a 10-year follow-up period, we examine relationships of long-term aspirin use with epigenetic mitotic clocks. We observed trends of mitotic clock IR deceleration in the proximal (cecum) colon samples of long-term aspirin users but not non-users.
Our rationale for conducting this analysis stemmed from the observation that many studies of epigenetic aging and CRC did not examine relationships with epigenetic mitotic clocks. This gap in biomarker utilization potentially stems from a lack of researcher familiarity. Both Hannum [19] and Horvath [10] epigenetic aging measures were first described in the literature in 2013 while MiAge [18], EpiTOC [16], and EpiTOC2 [17] were introduced in 2018, 2016, and 2020 respectively. Nevertheless, given the known relationships of mitosis with cancer [20], we hypothesized that the mitotic clocks may be more sensitive for detecting cancer relationships. Our data from this analysis of long-term aspirin use suggests that mitotic clocks may reflect similar physiology that is also captured by other epigenetic age markers.
In their original analysis of this data, Noreen et al. (2020) observed epigenetic age deceleration trends (via Hannum, Horvath, and PhenoAge metrics) in proximal but not distal colon samples [14]. Our observed, statistically significant and robust pattern of clock IR deceleration in the within group, long-term user analysis of proximal colon samples is consistent with their findings. Moreover, when comparing T2 minus T1 mitotic clock IR changes between non-users and long-term aspirin users, we observe more negative median IR differences in the proximal tissue of long-term users. Although not statistically significant, this is in line with prior observations of significant IR deceleration in the proximal tissue of long-term users at T2 compared to the proximal tissue of long-term users at T1. Unlike our within group comparisons, the between group, “difference,” comparisons were not paired. Thus, it is likely that our between group analyses did not meet statistical significance due to our sample size and a lack of statistical power. We hypothesized that analyses of differences within aspirin use groups are able to account for the within person variability yielding robust estimates.
Furthermore, given that the mitotic clocks demonstrated strong, positive associations with chronological age, we were not surprised to see increases in mitotic clock residuals not accounting for chronological age between T1 and T2 for each aspirin use group. These observations in addition to baseline differences in the ages of the two aspirin use groups (long-term users having a 10-year higher median chronological age than non-users) provide added impetus for focusing on the IR results—which take chronological age into account—for interpretation.
The fact that we observe significant IR deceleration in all three mitotic clocks suggests a good sensitivity of mitotic clocks to long-term aspirin relationships. However, in reviewing the literature, it remains unclear if sensitivity of mitotic clocks to CRC cancer processes is greater than that of other epigenetic clocks. The difference in robustness and sensitivity of epigenetic aging markers may be due to the amount of data used to create them. For instance, the Horvath metric was developed using approximately 8000 samples and the PhenoAge metric was developed using nearly 10,000 samples [10, 13]. In contrast, EpiTOC was developed using over 650 samples [16]. MiAge and EpiTOC2 were developed using approximately 4000 and 3000 samples respectively [17, 18]. Furthermore, we have observed this pattern in differential sensitivity to biological aging processes in an independent analysis. In a previous study of simulated long-duration space travel, we observed decreases in both PhenoAge and EpiTOC2, but only the PhenoAge deceleration remained statistically significant at the end of the 520-day simulated space travel mission [21]. In another study of healthy left colon samples from 334 individuals assigned to low, medium, or high risk groups based on personal history of adenomas or CRC, researchers observed significant PhenoAge deceleration in high risk individuals compared to low risk individuals; meanwhile, the relationships of Horvath, Hannum, and EpiTOC measures followed a similar trend but were not statistically significant [22]. Although these latter findings go against the paradigm of much of the CRC epigenetic aging literature—that higher cancer risk is associated with age acceleration—it provides added impetus for recommendations to help standardize future studies in an effort to reach a consensus on which epigenetic aging markers are best for studying CRC relationships. Consensus guidelines can also help facilitate meta-analyses which can further help in elucidating the true relationships and sensitivities of CRC-related biology with epigenetic clocks.
Though the data in this field is still in its nascency, we will reach a point where the field must establish a consensus for a biological aging gold standard [9]. Even if one general biological aging gold standard is not feasible, gold standards for particular domains or health outcomes (i.e., CRC) may be more plausible. As such, all studies within a particular domain will be expected to include certain standard markers even if more novel markers are the main focus of the study. Moving forward, we recommend that all epigenetic aging studies of CRC include traditional epigenetic aging markers (at minimum Hannum, Horvath, and PhenoAge) as well as mitotic clocks (at minimum MiAge, EpiTOC, and EpiTOC2 measures) to help the field reach a consensus on the already existing data. Likewise, the observation that epigenetic aging may differ based on colon location may be related to differential embryonic origins but is not fully understood [14, 23]. The pathways of tumor initiation are also known to differ between the right and left colon. Mutations in DNA mismatch repair pathways are more prevalent in right colon tumors whereas chromosomal instability pathway-related mutations are more prevalent in left colon tumors [23]. These distinct differences in molecular characteristics could have important relationships with epigenetic aging in CRC. Moreover, and potentially related to differences in embryologic origin and molecular characteristics, it is documented that some chemopreventive interventions like aspirin may be more robust in proximal colon versus distal colon [24]. Ultimately, additional research will be necessary to fully understand all of these relationships in the context of epigenetic aging. As such, we also recommend that epigenetic aging studies continue to provide colon location-specific results when possible. Finally, the racial disparities in colon cancer are well documented with African Americans suffering a greater CRC morbidity and mortality when compared to their peers [25]. A recent study reported age acceleration (Horvath metric) in the right colons of African Americans and deceleration in the right colons of their European American peers. The authors stated, “our results provide novel insight of epigenetic aging potentially underlying racial disparities in CRC [26].” We recommend that future studies of epigenetic aging in CRC—and otherwise—continue to be mindful of suggesting that disparities are solely due to ancestry. Devall et al. (2020) conducted a differential methylation position (DMP) analysis with race and found that 18.4% of DMPs overlapped with markers of ancestry; however, mentions of lifestyle and social factors as the potential drivers of their observed findings are lacking. We encourage researchers to outline and differentiate the role of racism and not ancestry among these biomarkers. This is particularly important given the reported associations of epigenetic aging with social factors including diet [13, 27], racial discrimination [28], neighborhood deprivation [29], and violence [30].
This study has some important limitations including its small sample size and limited generalizability given that the cohort was comprised of only Polish women. It is also critical to highlight the study’s observational design with no specifications on dose of aspirin taken during the follow-up period and limited covariates (we were only able to adjust for chronological age, BMI, polyp status, and methylation batch). Hence, we cannot directly speak to causality due to aspirin. Even in this specific instance where we are calculating metrics in colon tissue, associations of aspirin use with mitotic clocks and other epigenetic aging measures may be correlative and driven by other mechanisms such as immune system interactions. Future investigations in larger and more diverse populations as well as experimental studies will be necessary to confirm our findings more broadly. We believe that our proposed recommendations will also be important for improving the quality of these future studies.
Conclusion
Epigenetic aging measures, including mitotic clocks, demonstrate sensitivity for detecting CRC relationships. We specifically observed mitotic clock IR deceleration in the proximal colon mucosa of long-term aspirin users. Future studies that (1) include traditional epigenetic age as well as mitotic clock measures, (2) provide colon location-specific results, and (3) explore genetic as well as social drivers for racial epigenetic aging disparities will be critical for better characterizing epigenetic aging relationships in CRC.
Availability of data and materials
The dataset used in this study (Series GSE142257) is publicly available at: https://www.ncbi.nlm.nih.gov/geo.
Abbreviations
- CRC:
-
Colorectal cancer
- USPSTF:
-
United States Preventative Services Task Force
- EpiTOC2:
-
Epigenetic Time to Cancer-2
- EpiTOC:
-
Epigenetic Time to Cancer
- MiAge:
-
Mitotic age
- Timepoint 1/baseline:
-
T1
- Timepoint 2/10-year follow-up:
-
T2
References
Chan AT, Ladabaum U. Where do we stand with aspirin for the prevention of colorectal cancer? The USPSTF Recommendations. Gastroenterology. 2016;150(1):14–8. https://doi.org/10.1053/j.gastro.2015.11.018.
Katona BW, Weiss JM. Chemoprevention of colorectal cancer. Gastroenterology. 2020;158(2):368–88. https://doi.org/10.1053/j.gastro.2019.06.047.
Burn J, Sheth H, Elliott F, Reed L, Macrae F, Mecklin JP, et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: a double-blind, randomised, placebo-controlled trial. Lancet. 2020;395(10240):1855–63. https://doi.org/10.1016/S0140-6736(20)30366-4.
United States Preventive Services Taskforce Recommendation: Aspirin Use to Prevent Cardiovascular Disease and Colorectal Cancer: Preventive Medication. https://www.uspreventiveservicestaskforce.org/uspstf/recommendation/aspirin-to-prevent-cardiovascular-disease-and-cancer.
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. https://doi.org/10.5114/pg.2018.81072.
Hägg S, Jylhävä J. Should we invest in biological age predictors to treat colorectal cancer in older adults? Eur J Surg Oncol. 2020;46(3):316–20. https://doi.org/10.1016/j.ejso.2019.11.003.
Maugeri A, Barchitta M, Magnano San Lio R, Li Destri G, Agodi A, Basile G. Epigenetic aging and colorectal cancer: state of the art and perspectives for future research. Int J Mol Sci. 2020;22(1):200. https://doi.org/10.3390/ijms22010200.
Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–84. https://doi.org/10.1038/s41576-018-0004-3.
Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019;20(1):249. https://doi.org/10.1186/s13059-019-1824-y.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. https://doi.org/10.1186/gb-2013-14-10-r115.
Durso DF, Bacalini MG, Sala C, Pirazzini C, Marasco E, Bonafé M, et al. Acceleration of leukocytes’ epigenetic age as an early tumor and sex-specific marker of breast and colorectal cancer. Oncotarget. 2017;8(14):23237–45. https://doi.org/10.18632/oncotarget.15573.
Zheng C, Li L, Xu R. Association of epigenetic clock with consensus molecular subtypes and overall survival of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2019;28(10):1720–4. https://doi.org/10.1158/1055-9965.EPI-19-0208.
Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–91. https://doi.org/10.18632/aging.101414.
Noreen F, Chaber-Ciopinska A, Regula J, Schär P, Truninger K. Longitudinal analysis of healthy colon establishes aspirin as a suppressor of cancer-related epigenetic aging. Clinical Epigenetics. 2020;12(1):164. https://doi.org/10.1186/s13148-020-00956-9.
Boman BM, Fields JZ. An APC: WNT counter-current-like mechanism regulates cell division along the human colonic crypt axis: a mechanism that explains how APC mutations induce proliferative abnormalities that drive colon cancer development. Front Oncol. 2013;3:244. https://doi.org/10.3389/fonc.2013.00244.
Yang Z, Wong A, Kuh D, Paul DS, Rakyan VK, Leslie RD, et al. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biology. 2016;17(1):205. https://doi.org/10.1186/s13059-016-1064-3.
Teschendorff AE. A comparison of epigenetic mitotic-like clocks for cancer risk prediction. Genome Medicine. 2020;12(1):56. https://doi.org/10.1186/s13073-020-00752-3.
Youn A, Wang S. The MiAge Calculator: a DNA methylation-based mitotic age calculator of human tissue types. Epigenetics. 2018;13(2):192–206. https://doi.org/10.1080/15592294.2017.1389361.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda SV, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67. https://doi.org/10.1016/j.molcel.2012.10.016.
Levine MS, Holland AJ. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018;32(9-10):620–38. https://doi.org/10.1101/gad.314351.118.
Nwanaji-Enwerem JC, Nwanaji-Enwerem U, Van Der Laan L, et al. A longitudinal epigenetic aging and leukocyte analysis of simulated space travel: the Mars-500 mission. Cell Rep. 2020;33(10):108406. https://doi.org/10.1016/j.celrep.2020.108406.
Wang T, Maden SK, Luebeck GE, Li CI, Newcomb PA, Ulrich CM, et al. Dysfunctional epigenetic aging of the normal colon and colorectal cancer risk. Clin Epigenetics. 2020;12(1):5. https://doi.org/10.1186/s13148-019-0801-3.
Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterology Res. 2018;11(4):264–73. https://doi.org/10.14740/gr1062w.
Rothwell PM, Wilson M, Elwin C-E, Norrving B, Algra A, Warlow CP, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376(9754):1741–50. https://doi.org/10.1016/S0140-6736(10)61543-7.
Jackson CS, Oman M, Patel AM, Vega KJ. Health disparities in colorectal cancer among racial and ethnic minorities in the United States. J Gastrointest Oncol. 2016;7(Suppl 1):S32–43. https://doi.org/10.3978/j.issn.2078-6891.2015.039.
Devall M, Sun X, Yuan F, Cooper GS, Willis J, Weisenberger DJ, et al. Racial disparities in epigenetic aging of the right vs left colon. J Natl Cancer Inst. 2020. https://doi.org/10.1093/jnci/djaa206.
Phang M, Ross J, Raythatha JH, Dissanayake HU, McMullan RL, Kong Y, et al. Epigenetic aging in newborns: role of maternal diet. Am J Clin Nutr. 2020;111(3):555–61. https://doi.org/10.1093/ajcn/nqz326.
Brody GH, Miller GE, Yu T, Beach SRH, Chen E. Supportive family environments ameliorate the link between racial discrimination and epigenetic aging. Psychol Sci. 2016;27(4):530–41. https://doi.org/10.1177/0956797615626703.
Lawrence KG, Kresovich JK, O’Brien KM, Hoang TT, Xu Z, Taylor JA, et al. Association of neighborhood deprivation with epigenetic aging using 4 clock metrics. JAMA Netw Open. 2020;3(11):e2024329. https://doi.org/10.1001/jamanetworkopen.2020.24329.
Jovanovic T, Vance LA, Cross D, Knight AK, Kilaru V, Michopoulos V, et al. Exposure to violence accelerates epigenetic aging in children. Sci Rep. 2017;7(1):8962. https://doi.org/10.1038/s41598-017-09235-9.
Acknowledgements
The authors acknowledge and thank Faiza Noreen, Anna Chaber-Ciopinska, Jaroslaw Regula, Primo Schär, and Kaspar Truninger for making their dataset publicly available.
Funding
JNE and AC are supported by the National Institutes of Health grants R03AG067064 and R01ES031259.
Author information
Authors and Affiliations
Contributions
JCN conceived and designed the study. JCN performed the data analyses. JCN and AC drafted the manuscript. All authors contributed to the analyses and revised and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was an analysis of publicly available data.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table S1.
Baseline Versus 10-Year Follow-Up Within Group Comparisons of Median Mitotic Clock Residuals for Non-Users and Long-term Aspirin Users. Supplementary Table S2. Baseline Versus 10-Year Follow-Up Comparisons of Median Mitotic Clock Residual Differences between Non-Users and Long-term Aspirin Users.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nwanaji-Enwerem, J.C., Nze, C. & Cardenas, A. Long-term aspirin use and epigenetic mitotic clocks for cancer risk prediction: findings in healthy colon mucosa and recommendations for future epigenetic aging studies. Epigenetics Commun. 1, 5 (2021). https://doi.org/10.1186/s43682-021-00004-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43682-021-00004-4