Abstract
Background
Repeated attacks of acute exacerbations of chronic obstructive pulmonary disease (AECOPD) have been attributed to bacterial infections. However, sputum microbiology for identifying the bacteriological profile is not performed usually and remains controversial. Thus, we performed a prospective, cross-sectional study to assess the clinico-bacteriological profile of the patients with AECOPD and antibiotic resistance pattern of bacteria isolated from sputum samples. The study involved 104 consecutive hospitalized patients, of either gender, aged 40–90 years, and diagnosed with AECOPD. Before initiating an empirical antibiotic therapy, the sputum samples of all the patients were collected and subjected to Gram staining, bacterial culture, and antibiotic sensitivity.
Results
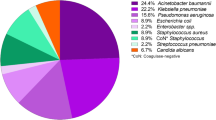
The most common bacterial isolates were P. aeruginosa (30.7%) followed by K. pneumoniae (20.3%) and S. pneumoniae (8.6%). Both Gram-positive and Gram-negative bacteria were most sensitive to Meropenem (91%) followed by Amoxicillin-clavulanic acid (83%). However, these bacteria were most resistant to Cefoperazone-sulbactam (43%) followed by Levofloxacin (41%). P. aeruginosa and K. pneumoniae were most resistant to Cefoperazone-sulbactam (23%) followed by Fluoroquinolones (23–35%), while S. pneumoniae was most resistant to Ciprofloxacin (44%) followed by Amoxicillin-clavulanic acid (33%).
Conclusions
Isolation of P. aeruginosa, as the most common agent, further confirms its roles in severe attacks of AECOPD. The regularly used antibiotics were found to be resistant to most isolates, thereby suggesting that severity of AECOPD may be attributed to the prevalence of antibiotic-resistant strains.
Similar content being viewed by others
Background
Chronic obstructive pulmonary disease (COPD), though preventable, is an incapacitating and progressive airway disease of multifactorial origin [1, 2]. Acute exacerbation of COPD (AECOPD) is marked by acute aggravation of respiratory symptoms that necessitate alteration in the treatment [3]. These persistent attacks result in ferocious decrease in lung function and patients with frequent attacks of AECOPD have a significantly reduced 5-year survival, especially if they require hospital care [4].
Respiratory tract infections (RTIs) are widely recognized as the most frequent precipitating factor [5, 6]. The commonly associated bacteria include Haemophilus influenza, Moraxella catarrhalis, Streptococcus pneumonia, Pseudomonas, and others [7]. Currently, antibiotics are recommended for the treatment of AECOPD in the presence of bacterial infection [3]. However, antibiotics are chosen on the basis of the locally prevalent bacteria and their resistance pattern [8].
The use of sputum cultures to reach a diagnosis and aid the clinical management of AECOPD are controversial. In view of the above, along with the limited available data, the present study was carried out to assess the clinico-bacteriological profile of patients with AECOPD and antibiotic resistance pattern of bacteria isolated from sputum samples.
Methods
The present study was hospital-based and cross-sectional in nature. It included 104 consecutive patients with AECOPD, admitted in the Department of Pulmonary Medicine, ESIC Medical College, Hyderabad, India, over a period of 21 months (January 2018 to September 2019). The study commenced after obtaining approval from Institutional Ethics Committee and written informed consent of the patients.
The study included patients of either gender, aged 40–90 years, diagnosed with AECOPD (as per the Global Initiative for Chronic Obstructive Lung Disease [GOLD] 2018 criteria for the diagnosis) [9], requiring in-patient admission in wards and adequate sputum sample (<10 squamous epithelial cells and >25 pus cells), while the patients diagnosed with tuberculosis, bronchial asthma, bronchiectasis, malignancy, and community acquired pneumonia; those who received antibiotic therapy in the past 21 days; those managed in out-patient or emergency department; those admitted in in-patient department or intensive care unit; and those with ischemic heart disease were excluded from the study.
Study procedure
The patient fulfilling the eligibility criteria were included in the study. Following enrolment, in all the patients, detailed history was noted and both general and systemic examinations were performed. Variables such as age, gender, presenting symptoms, history of smoking, duration of smoking, and smoking index were recorded.
Following clinical examination, each patient underwent complete blood counts, differential blood counts, chest radiography, and pulse oximetry. This was followed by spirometry and collection of sputum samples.
Spirometry
In all the patients, spirometry (Zan 300, Sensor Medics MGA USB, Oberthulba, Germany) was performed to determine the severity of airflow limitation. The post-bronchodilator ratio of forced expiratory volume in one second to forced vital capacity (FEV1/FVC) < 0.70 was considered for the clinical diagnosis of COPD. Moreover, as per GOLD 2018 guidelines, in patients with COPD, the post-bronchodilator FEV1 ≥ 80% predicted, 50 ≤ FEV1 < 80% predicted, 30 ≤ FEV1 < 50% predicted, and FEV1< 30% predicted were considered as mild, moderate, severe, and very severe airway limitation [9].
Sputum sample collection and culture
In all patients, based on the standard guideline, early morning deep coughed sputum sample was collected [10]. Within 24-h post-admission, patients were instructed to collect sputum into a sterile container with a screw cap. Prior to sputum collection, to avoid oral contamination of the sample, patients were asked to rinse the mouth twice with water and antiseptic solution. Within the next 2 h, the samples were transported to and processed in the Microbiology Department. The samples were then subjected to Gram staining and culture on two sheep blood agar, chocolate agar, and McConkey agar plates. One sheep blood agar streaking with staphylococci was performed to aid H. influenzae growth. All plates were incubated for 24–48 h at 37°C, while sheep blood agar plates were kept in 5–10% CO2 incubator. Gram stain was reported on the basis of Bartlett’s grading system [11], and culture isolates were identified by standard techniques [12]. Based on the standards laid down by the Clinical Laboratory Standards Institute, Kirby-Bauer method was used to test antibiotic sensitivity of the pathogenic organisms isolated in culture [13].
Results
The majority of the patients were male (79.8%). The mean age of the study population was 66.8 ± 11.4 years. Distribution of patients according to the age groups revealed a sequential rise in the number of patients from 40–49 years (8.7%) to reach the maximum in 70–79 years (32.6%), only to drop in the 80–89 years (14.4%) age group. The most common presenting symptom was shortness of breath (98.1%) followed by cough (90.4%) and expectoration (89.4%). On spirometry, majority of the patients were found to have severe airway limitation (54.8%) (Table 1).
Evaluation of smoking as a risk factor revealed that 85 (81.7%) patients were smokers and among them 78 (93.9%) were males. On enquiry, majority of the patients had a history of smoking for the past 20–29 years (35.3%) followed by 30–39 years (29.4%). Similar pattern was observed in males, i.e., 37.2% and 29.5%, respectively, while majority of the females indulged in smoking for 10–19 years (57.1%) followed by 30–39 years (28.6%). The mean smoking index was 219.7 ± 148.8. Majority of the patients had a smoking index of 100–300 (56.5%) followed by < 100 (25.9%). Similar pattern was observed in males, i.e., 60.3% followed by 23.1%, respectively, while majority of the females had a smoking index of < 100 (57.1%) followed by > 300 (28.6%) (Table 2).
On bacteriological culture, most commonly grown bacterial isolates were P. aeruginosa (30.7%) followed by Klebsiella pneumoniae (20.3%), S. pneumoniae (8.6%), and H. influenzae (7.7%), while the least commonly observed isolates were Acinetobacter (1.9%) followed by M. catarrhalis (3.8%), S. aureus (4.8%), and S. pyogenes (4.8%) (Table 3).
Examination of antibiotic sensitivity pattern revealed that both Gram-positive and Gram-negative bacteria were most sensitive to Meropenem (91%) followed by Amoxicillin-clavulanic acid (83%) and Ceftriaxone (63%), while these bacteria were most resistant to Cefoperazone-sulbactam (43%) followed by Levofloxacin (41%) and Amikacin (41%) (Table 4).
Examination of antibiotic resistance pattern among the most common isolates revealed that P. aeruginosa was most resistant to Cefoperazone-sulbactam (23%) followed by Ciprofloxacin (23%). Similarly, K. pneumoniae was most resistant to Cefoperazone-sulbactam (55%) followed by Levofloxacin (35%), while S. pneumoniae was most resistant to Ciprofloxacin (44%) followed by Amoxicillin-clavulanic acid (33%) (Table 5).
Discussion
Repeated attacks of AECOPD adversely affect the patients resulting in hospitalization, readmission, progression of disease, and death [4]. The odds of developing an attack of exacerbation depends on the agent involved and are 1.69, 1.77, and 2.96 for H. influenzae, S. pneumoniae, and M. catarrhalis, respectively [14]. Though agents such as H. influenzae and S. pneumonia have been isolated in patients with stable COPD, relatively higher loads of these isolates have been found during exacerbations [15]. According to a study, the most prevalent species isolated during exacerbation were S. pneumoniae followed by H. influenzae, M. catarrhalis, and Legionella pneumophila, while in patients with more severe COPD, P. aeruginosa was also commonly implicated in the causation of exacerbations [16]. Moreover, compared with exacerbations not associated with P. aeruginosa, its presence suggests a grave prognosis with greater 30- and 90-day death rates [17].
In the present study, P. aeruginosa, K. pneumoniae, S. pneumoniae, and H. influenzae, in the decreasing order, were the most common bacterial isolates. Moreover, majority of the patients were found to have severe and very severe airway limitation (65.4%) and required hospitalization. Thus, presence of P. aeruginosa as the most common isolate can be attribute to the severity of the disease. Similar to the present study, Chawla et al. [18] and Shah et al. [19] reported P. aeruginosa as the most common isolate in the hospitalized patients. However, studies by Sharan [20] and Jog et al. [21] reported K. pneumoniae as the most common isolate. Other studies by Sharma et al. [8] and Shashibhushan et al. [22] reported S. pneumoniae as the most common isolate. The association of P. aeruginosa with severity of COPD has already been documented. A recent study by Jacobs et al. concluded that isolates of P. aeruginosa not only acts as a marker of worsening disease but suggests adverse clinical outcomes. They reported that presence of P. aeruginosa was associated with significantly greater risk of mortality and higher rates of exacerbation and hospitalization [23]. Another study by Eklof et al. reported similar findings [24].
A recent study reported that patients with microorganism resistant to regularly used antibiotics had higher episodes of AECOPD in the past 1 year, more severe disease, higher dyspnea, and a positive respiratory culture in the last year (particularly for P. aeruginosa); received more antibiotics in the last 3 months and had longer hospital stay [25]. Moreover, past studies involving patients with severe AECOPD reported that this sub-group of population have an increased frequency of exacerbations, prior antibiotic use, history of hospitalization, and more severe airflow limitations [26,27,28]. Thus, the strategy of choosing an antibiotic, based on the resistance pattern, is necessary for both the efficient and economical management and decreasing the chances of development of drug resistance [8].
In the present study, sensitivity testing revealed a significant resistance against commonly used antibiotics and the resistance ranged from 17 to 43%. Both Gram-positive and Gram-negative bacteria were most resistant to Cefoperazone-sulbactam followed by Levofloxacin and Amikacin. The meticulous and judicious use of antibiotics based on the stringent antimicrobial use policy of the hospital may explain the relatively low antimicrobial resistance rate in our study. Isolates were most sensitive to Meropenem followed by Amoxicillin-clavulanic acid and Ceftriaxone. In a study by Hassan et al., bacterial resistance rates were the highest against Amoxicillin, Amoxicillin-clavulanic acid, and Cephalosporins; moderate against Fluoroquinolones (ofloxacin, levofloxacin, ciprofloxacin); and least against Carbapenems [29]. In the present study, around 40% isolates were resistant to both ciprofloxacin and levofloxacin. Similar finding was reported by Hassan et al., where about 50% isolates were resistant to both ciprofloxacin and levofloxacin [29]. Madhavi et al. reported a similar rate of sensitivity to ciprofloxacin [30]. A study by Lode et al. reported high bacteriological eradication rate with levofloxacin [31]. Thereby, suggesting the sensitivity towards levofloxacin.
In the present study, sensitivity testing revealed that P. aeruginosa and K. pneumoniae were most resistant to Cefoperazone-sulbactam followed by Fluoroquinolones (Ciprofloxacin and Levofloxacin). However, S. pneumoniae was most resistant to Ciprofloxacin followed by Amoxicillin-clavulanic acid. Though P. aeruginosa and S. pneumoniae were completely susceptible to Meropenem, K. pneumoniae had slight resistance. Moreover, both P. aeruginosa and S. pneumoniae had least resistance against Piperacillin-tazobactam. Similarly, Shashibhushan et al. reported that P. aeruginosa was sensitive to Piperacilin-tazobactum, Amikacin, and Levofloxacin. In the same study, Ceftriaxone was most effective against K. pneumoniae and S. pneumoniae [22]. In another study, Aleemullah et al. reported that P. aeruginosa was highly sensitive to Imipenem, Piperacillin-Tazobactam, and Amikacin [32]. Hassan et al. reported that K. pneumoniae, P. aeruginosa, and M. catarrhalis were completely susceptible to the Carbapenem antibiotics [29]. Patel et al. reported Piperacillin-tazobactam as most effective against S. pneumonia [33]. In their study, Sharma et al. reported that S. pneumoniae was sensitive to commonly prescribed antibiotics such as Macrolides, Cephalosporins, and Quinolones [8].
Amongst various known risk factors of AECOPD, important ones include RTIs, smoking, air pollutants, severe limitation in airflow, and old age. The prevalence of COPD increases clearly in individuals aged ≥ 40 years [5]. This can be attributed to the fact that chronic bronchitis has highest prevalence in fifth and sixth decade of life [34]. In the present study, the mean age of the patients was 66.8 ± 11.4 years and maximum prevalence was observed in the age group of 70–79 years. Thus, AECOPD is frequently observed in patients belonging to advanced age group. In this population, the airway is more prone to exacerbation, as a result of impaired immunological defenses, associated co-morbidities, increased duration of seasonal variation, tobacco smoke, and environmental pollutants.
In the present study, males comprised the majority, with male to female ratio of 3.9. Majority of the patients were smokers, with males outnumbering the females. Among the females, only one-third indulged in smoking. The predominant prevalence of COPD in males can be attributed to the fact that males are more mobile and involved in outdoor activities than females and thus are subjected to more environmental pollutants. Moreover, males tend to indulge more in smoking and smoking is recognized as a risk factor for COPD and precipitation of exacerbation. Smoking and air pollution are known to decrease mucociliary clearance and alter innate immunity. This leads to increased colonization of bacteria and other pathogens in lower airway that results in rise of airway inflammation and precipitates an attack of exacerbation [35]. In non-smokers, particularly among women, development of COPD is explained by exposure to indoor air pollution [36].
Various studies have reported a relationship between AECOPD and the degree of airflow limitation. Mullerova et al. reported that the odds of frequent attacks of exacerbations increases with increased severity of airway limitation, and it is 1.2 and 2.4 for moderate and very severe limitation, respectively [37]. Another study by Au et al. demonstrated that rate of hospitalization increases with increase in airway limitation [38]. Montes de Oca et al. highlighted that severity of airway limitation and dyspnea were directly correlated with the rise in hospital visits [39]. Similarly, in the present study, majority of the patients were found to have severe and very severe airway limitation. Moreover, the most common presenting symptom was shortness of breath followed by cough and expectoration, thereby suggesting coexistence of airway resistance and inflammation in patients with severe attacks of exacerbation.
Conclusions
In conclusion, P. aeruginosa and K. pneumoniae were the most common bacterial isolates. The bacteriological profile of the sputum samples suggests the role of commonly encountered pathogens in the precipitation of AECOPD and highlights the differences in causative agents, with respect to other studies. Isolation of P. aeruginosa, as the most common agent, further confirms its roles in severe attacks of AECOPD. The regularly used antibiotics were found to be ineffective against most isolates, thereby suggesting that severe nature of AECOPD may be attributed to the prevalence of antibiotic-resistant strains. The study further highlights the sensitivity of broad spectrum carbapenems against both Gram-positive and Gram-negative isolates. The bacteriological profile in the present study is of concern to the emerging bacterial population, their resistance patterns, and warrants regular (or annual) local studies to update our local antibiogram and antimicrobial resistance pattern, and accordingly change our empiric antimicrobial therapy.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AECOPD:
-
Acute exacerbation of chronic obstructive pulmonary disease
- COPD:
-
Chronic obstructive pulmonary disease
- FEV1:
-
Forced expiratory volume in one second
- FVC:
-
Forced vital capacity
- RTI:
-
Respiratory tract infection
References
Agusti A, Hogg JC (2019) Update on the pathogenesis of chronic obstructive pulmonary disease. N Engl J Med 381(13):1248–1256 https://doi.org/10.1056/NEJMra1900475
Lee J, Jung HM, Kim SK, Yoo KH, Jung KS, Lee SH et al (2019) Factors associated with chronic obstructive pulmonary disease exacerbation, based on big data analysis. Sci Rep 9(1):6679. https://doi.org/10.1038/s41598-019-43167-w
Global Initiative for Chronic Obstructive Lung Disease. Edition 2020. Available at: https://goldcopd.org/wp-content/uploads/2020/03/GOLD-2020-POCKET-GUIDE-ver1.0_FINAL-WMV.pdf [Accessed on 14 Mar 2021].
Soler-Cataluna JJ, Martinez-Garcia MA, Roman Sanchez P, Salcedo E, Navarro M, Ochando R (2005) Severe acute exacerbations and mortality in patients with chronic obstructive pulmonary disease. Thorax 60(11):925–931. https://doi.org/10.1136/thx.2005.040527
Hogea SP, Tudorache E, Fildan AP, Fira-Mladinescu O, Marc M, Oancea C (2020) Risk factors of chronic obstructive pulmonary disease exacerbations. Clin Respir J 14(3):183–197. https://doi.org/10.1111/crj.13129
Miravitlles M, Anzueto A (2015) Role of infection in exacerbations of chronic obstructive pulmonary disease. Curr Opin Pulm Med 21(3):278–283. https://doi.org/10.1097/MCP.0000000000000154
Miravitlles M, Anzueto A (2017) Chronic respiratory infection in patients with chronic obstructive pulmonary disease: what is the role of antibiotics? Int J Mol Sci 18(7):1344. https://doi.org/10.3390/ijms18071344
Sharma P, Narula S, Sharma K, Kumar N, Lohchab K, Kumar N (2017) Sputum bacteriology and antibiotic sensitivity pattern in COPD exacerbation in India. Egypt J Chest Dis Tuberc 66(4):593–597. https://doi.org/10.1016/j.ejcdt.2017.08.003
Global Initiative for Chronic Obstructive Lung Disease. Edition 2018. Available at: https://goldcopd.org/wp-content/uploads/2017/11/GOLD-2018-v6.0-FINAL-revised-20-Nov_WMS.pdf [Accessed on 14 Mar 2021].
Baron EJ, Thomson JR (2012) Specimen collection, transport and processing: bacteriology. In: Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW (eds) Manual of clinical microbiology, 10th edn. ASM Press, Washington, DC., pp 228–271
Winn WC, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, Woods GL (2006) Koneman’s colour atlas and textbook of diagnostic microbiology Chapter 1, 6th edn. Lippincott Williams and Wilkins, Philadelphia, pp 1–66
Barrow GI, Feltham RKA (1993) Cowan and Steel’s manual for the identification of medical bacteria; 3rd edn. Cambridge: Cambridge University Press
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. M100, 28th ed. January 2018. Available at: https://file.qums.ac.ir/repository/mmrc/CLSI-2018-M100-S28.pdf [Accessed on 24 Nov 2021].
Leung JM, Tiew PY, Mac Aogáin M, Budden KF, Yong VF, Thomas SS et al (2017) The role of acute and chronic respiratory colonization and infections in the pathogenesis of COPD. Respirology 22(4):634–650. https://doi.org/10.1111/resp.13032
Garcha DS, Thurston SJ, Patel AR, Mackay AJ, Goldring JJ, Donaldson GC et al (2012) Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 67(12):1075–1080. https://doi.org/10.1136/thoraxjnl-2012-201924
Domenech A, Puig C, Marti S, Santos S, Fernandez A, Calatayud L et al (2013) Infectious etiology of acute exacerbations in severe COPD patients. J Infect 67(6):516–523. https://doi.org/10.1016/j.jinf.2013.09.003
Rodrigo-Troyano A, Suarez-Cuartin G, Peiro M, Barril S, Castillo D, Sanchez-Reus F et al (2016) Pseudomonas aeruginosa resistance patterns and clinical outcomes in hospitalized exacerbations of COPD. Respirology 21(7):1235–1242 https://doi.org/10.1111/resp.12825
Chawla K, Mukhopadhay C, Majumdar M, Bairy I (2008) Bacteriological profile and their antibiogram from cases of acute exacerbations of chronic obstructive pulmonary disease: a hospital based study. J Clin Diagn Res 2(1):612–616 http://eprints.manipal.edu/id/eprint/70
Shah BA, Singh G, Naik MA, Dhobi GN (2010) Bacteriological and clinical profile of community acquired pneumonia in hospitalized patients. Lung India 27(2):54–57 https://doi.org/10.4103/0970-2113.63606
Sharan H (2015) Aerobic bacteriological study of acute exacerbations of chronic obstructive pulmonary disease. J Clin Diagn Res 9(8):DC10–DC12 https://doi.org/10.7860/JCDR/2015/14515.6367
Jog AP, Davita TR (2018) Bacteriological profile of patients with acute exacerbation of COPD. Int J Adv Res 6(1):348–355. https://doi.org/10.21474/IJAR01/6225
Shashibhushan BL, Nagaraja C, Arun BJ, Nagaraj N (2016) Bacteriological profile and antibiotic sensitivity pattern in sputum culture of chronic obstructive pulmonary disease patients. Int J Adv Med 3(3):671–674. https://doi.org/10.18203/2349-3933.ijam20162515
Jacobs DM, Ochs-Balcom HM, Noyes K, Zhao J, Leung WY, Pu CY et al (2020) Impact of Pseudomonas aeruginosa isolation on mortality and outcomes in an outpatient chronic obstructive pulmonary disease cohort. Open Forum Infect Dis 7(1):ofz546. https://doi.org/10.1093/ofid/ofz546
Eklof J, Sørensen R, Ingebrigtsen TS, Sivapalan P, Achir I, Boel JB et al (2020) Pseudomonas aeruginosa and risk of death and exacerbations in patients with chronic obstructive pulmonary disease: an observational cohort study of 22,053 patients. Clin Microbiol Infect 26(2):227–234. https://doi.org/10.1016/j.cmi.2019.06.011
Estirado C, Ceccato A, Guerrero M, Huerta A, Cilloniz C, Vilaro O et al (2018) Microorganisms resistant to conventional antimicrobials in acute exacerbations of chronic obstructive pulmonary disease. Respir Res 19:119. https://doi.org/10.1186/s12931-018-0820-1
Soler N, Torres A, Ewig S, Gonzalez J, Celis R, El-Ebiary M et al (1998) Bronchial microbial patterns in severe exacerbations of chronic obstructive pulmonary disease (COPD) requiring mechanical ventilation. Am J Respir Crit Care Med 157(5 Pt 1):1498–1505. https://doi.org/10.1164/ajrccm.157.5.9711044
Miravitlles M, Espinosa C, Fernández-Laso E, Martos JA, Maldonado JA, Gallego M (1999) Relationship between bacterial flora in sputum and functional impairment in patients with acute exacerbations of COPD. Study Group of Bacterial Infection in COPD. Chest 116(1):40–46. https://doi.org/10.1378/chest.116.1.40
Eller J, Ede A, Schaberg T, Niederman MS, Mauch H, Lode H (1998) Infective exacerbations of chronic bronchitis: relation between bacteriologic etiology and lung function. Chest 113(6):1542–1548. https://doi.org/10.1378/chest.113.6.1542
Hassan AT, Mohamed SAA, Mohamed MSE, El-Mokhtar MA (2016) Acute exacerbations of chronic obstructive pulmonary disease: etiological bacterial pathogens and antibiotic resistance in Upper Egypt. Egypt J Bronchol 10:283–290 https://doi.org/10.4103/1687-8426.193640
Madhavi S, Rama Rao MV, Janardhan Rao R (2012) Bacterial etiology of acute exacerbations of chronic obstructive pulmonary disease. J Microbiol Biotech Res 2(3):440–444 http://scholarsresearchlibrary.com/JMB-vol2-iss3/JMB-2012-2-3-440-444.pdf
Lode H, Eller J, Linnhoff A, Ioanas M (2004) Evaluation of Therapy-Free Interval in COPD Patients Study Group. Levofloxacin versus clarithromycin in COPD exacerbation: focus on exacerbation-free interval. Eur Respir J 24(6):947–953. https://doi.org/10.1183/09031936.04.00009604
Aleemullah MF, Krishnamurthy V, Harish M, Akeel CA (2016) Bacteriological profile of patients with AECOPD-hospital based study. Int J Curr Microbiol App Sci 5(4):84–90. https://doi.org/10.20546/ijcmas.2016.504.012
Patel AK, Luhadia AS, Luhadia SK (2015) Sputum bacteriology and antibiotic sensitivity pattern of patients having acute exacerbation of COPD in India – a preliminary study. J Pulm Respir Med 5(1):238. https://doi.org/10.4172/2161-105X.1000238
Mejza F, Gnatiuc L, Buist AS et al (2017) Prevalence and burden of chronic bronchitis symptoms: results from the BOLD study. Eur Respir J 50(5):1700621. https://doi.org/10.1183/13993003.00621-2017
Strzelak A, Ratajczak A, Adamiec A, Feleszko W (2018) Tobacco smoke induces and alters immune responses in the lung triggering inflammation, allergy, asthma and other lung diseases: a mechanistic review. Int J Environ Res Public Health 15(5):1033. https://doi.org/10.3390/ijerph15051033
Rajendra KC, Shukla SD, Gautam SS, Hansbro PM, O'Toole RF (2018) The role of environmental exposure to non-cigarette smoke in lung disease. Clin Transl Med 7(1):39. https://doi.org/10.1186/s40169-018-0217-2
Müllerová H, Shukla A, Hawkins A, Quint J (2014) Risk factors for acute exacerbations of COPD in a primary care population: a retrospective observational cohort study. BMJ Open 4(12):e006171. https://doi.org/10.1136/bmjopen-2014-006171
Au LH, Chan HS (2013) Severity of airflow limitation, co-morbidities and management of chronic obstructive pulmonary disease patients acutely admitted to hospital. Hong Kong Med J 19(6):498–503. https://doi.org/10.12809/hkmj133909
Montes de Oca MM, Aguirre C, Varela MVL, Laucho-Contreras ME, Casas A, Surmont F (2016) Exacerbations and health care resource utilization in patients with airflow limitation diseases attending a primary care setting: the PUMA study. Int J Chron Obstruct Pulmon Dis 11:3059–3067. https://doi.org/10.2147/COPD.S120776
Acknowledgements
The authors would like to thank Dr. Vikas S. Sharma (MD), Principal Consultant, Maverick MedicorumTM (India), for medical writing assistance in the preparation of this article.
Funding
Authors received no funding for this study.
Author information
Authors and Affiliations
Contributions
NM and AKB were involved in study design, data collection, data interpretation, analysis of results, and manuscript writing. SRK and JC were involved in study design, data interpretation, analysis of results, and manuscript writing. The authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was done after due permission from the Institutional Ethics Committee and after taking written informed consent from the patients. The study was approved by the Institutional Ethics Committee, Employee’s State Insurance Corporation Medical College vide reference no. 799/U/IEC/ESICMC/F0108/04-2019 dated 15/04/2019.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mood, N., Katta, S.R., Badam, A.K. et al. Clinico-bacteriological profile and antibiotic resistance pattern in patients with acute exacerbation of COPD. Egypt J Intern Med 34, 13 (2022). https://doi.org/10.1186/s43162-021-00094-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43162-021-00094-5