Abstract
Background
Vibrio species are among the autochthonous bacterial populations found in surface waters and associated with various life-threatening extraintestinal diseases, especially in human populations with underlying illnesses and wound infections. Presently, very diminutive information exists regarding these species’ mutational diversity of virulence and resistance genes. This study evaluated variations in endonucleases and mutational diversity of the virulence and resistance genes of Vibrio isolates, harboring virulence-correlated gene (vcgCPI), dihydropteroate synthase type 1 and type II genes (Sul 1 and 11), (aadA) aminoglycoside (3′′) (9) adenylyltransferase gene, (aac(3)-IIa, (aacC2)a, aminoglycoside N(3)-acetyltransferase III, and (strA) aminoglycoside 3′-phosphotransferase resistance genes.
Methods
Using combinations of molecular biology techniques, bioinformatics tools, and sequence analysis.
Results
Our result revealed various nucleotide variations in virulence determinants of V. vulnificus (vcgCPI) at nucleotide positions (codon) 73–75 (A → G) and 300–302 (N → S). The aminoglycosides resistance gene (aadA) of Vibrio species depicts a nucleotide difference at position 482 (A → G), while the aminoglycosides resistance gene (sul 1 and 11) showed two variable regions of nucleotide polymorphism (102 and 140). The amino acid differences exist with the nucleotide polymorphism at position 140 (A → E). The banding patterns produced by the restriction enzymes HinP1I, MwoI, and StyD4I showed significant variations. Also, the restriction enzyme digestion of protein dihydropteroate synthase type 1 and type II genes (Sul 1 and 11) differed significantly, while enzymes DpnI and Hinf1 indicate no significant differences. The restriction enzyme NlaIV showed no band compared to reference isolates from the GenBank. However, the resistant determinants show significant point nucleotide mutation, which does not produce any amino acid change with diverse polymorphic regions, as revealed in the restriction digest profile.
Conclusion
The described virulence and resistance determinants possess specific polymorphic locus relevant to pathogenomics studies, pharmacogenomic, and control of such water-associated strains.
Similar content being viewed by others
Background
Most Vibrio species are human pathogens [1, 2] and disease-causing strains that have been particularly implicated in gastroenteritis and the infection of open wounds, causing sepsis [3]. These species are primarily present in water and food and carried by many marine animals, such as crabs or prawns, which carry the bacteria that can cause fatal illnesses if exposed [4,5,6,7].
Several genomic, proteomic, and genetic markers have been applied to the pathogenic profile of the water-loving Vibrio species [6]. In particular, primary pathogenic/epidemic genetic markers/genes for V. cholerae include ctxAB, tcpA, hap, and toxR, which codes for cholera toxin, toxin-coregulated adhesion pili, soluble hemagglutinin/protease, and regulatory toxoid [2, 8, 9]. While V. parahaemolyticus has the genetic marker O3:K6 antigens that regulate the serovar, also the genes toxRS [10], orf8 [11], and tdh; and trh, found in most of the pathogenic strains. The V. Vulnificus markers involve pathogenicity region XII, nanA, and a mannitol fermentation operon containing alleles of the 16S rRNA and vcg genes linked with pathogenicity [12]. V. mimicus genetic factors include; quorum-sensing regulation system, hemolysins, proteases, outer membrane proteins [(OmpU), OmpT, OmpK, and OmpV] [2], a type IV and MSHA pilus, an aerobactin siderophore, a capsular polysaccharide, an accessory colonization factor (acfD), the transmembrane regulatory protein ToxS, the transcriptional activator ToxR, and the presence of quorum- (LuxS, LuxO, LuxR) [13]. While other pathogenic vibriosis shares common and/or combined genetic markers. It is imperative to note that some Vibrio spp., show no positive result to the aforementioned genetic markers but are potential pathogens, implying the discrimination markers insufficient to trace the toxins in the bacterial isolate in environment samples [7].
In addition, multiple drug resistance is well reported among the Vibrio strains highlighting mechanisms via resistance coding genes [9], the acquisition of conjugative plasmids [14,15,16], genetic elements (class 1 integron and SXT elements), a potential carrier of antimicrobial resistance genetic determinants [9, 17, 18]. Also, conjugative elements (ICEs) are a type of mobile genetic element that encodes various characteristics, including drug resistance [19]. Specifically, the SXT element helps horizontal resistance gene transfer and rearrange resistance genes in V. cholerae. It was initially found in the V. cholerae O139 MO10 chromosome from India (SXTMO10) but was later observed in other strains [20]. This element can mobilize plasmids, integron genes, and other resistant genes, including chloramphenicol (coded by floR), streptomycin (strA and strB), sulfamethoxazole (sul1 and sul2), trimethoprim (dfrA18), Penicillins (AmpC), lactamase for Cephalosporins, (blaSHV, blaTEM, blaCTX-M) Carbapenems (blaNDM-1, blaKPC, blaIMP, blaVIM), Macrolides (vanA, mecA), and Fluoroquinolones (mcr-1) and tetracycline (tetA gene) [7, 21,22,23,24]. High levels of resistance to sulfamethoxazole (sul2), chloramphenicol (floR), streptomycin (strA and strB), and trimethoprim (dfrA1) have been documented [18], which are associated with the integrase gene, SXT int, and associated SXT resistance genes. At the same time, there are variant types of the SXT element among pathogenic Vibrio spp. (Vibrio vulnificus, Vibrio metschnikovii, Vibrio fluvialis, and Vibrio parahaemolyticus) harbor these resistance genes [2, 25, 26].
Understanding the wide variations or mutations in virulence and resistance genes, including genetic and pathogenic diversity in natural environments among Vibrio species, are important and relevant indices for control, especially among other strains of Vibrio. Like other infectious diseases, typically fluoroquinolone resistance has been attributed to amino acid changes at positions Ser79 of ParC and Ser81 of GyrA to either Phe or Tyr (8, 33) [27, 28]. However, the appropriate codons' single-base modifications cannot account for these amino acid alterations, often they are second-step substitutions caused by 2-bp changes to the serine codons at ParC (TCT to CTT) or GyrA (TCC to ATC), respectively [27]. Also, mutations detected in the QRDRs of GyrA (Ser83-Ile) and ParC (Ser85-Leu) revealed the mechanisms for nalidixic acid resistance among Vibrio strains [26]. These mutations of a set of mobile fluoroquinolone resistance genes (qnr-genes), are implicated in the contamination of microbial communities. For instance, the chromosomal resistance mutations can arise de novo and become abundant in a population with strong sufficient antibiotic selective pressure, thereby confirming clinically relevant resistance. However, the abundance and distributions of these chromosomal resistance mutations in environmental bacterial communities are poorly investigated.
Pulsed-field gel electrophoresis (PFGE) uses appropriate restriction enzymes to break down bacterial DNA at a select few locations in the genome, resulting in big or macro-DNA fragments that may be sorted based on size. It has been demonstrated that PFGE banding patterns produced by NotI restriction are a useful genotypic tool for identifying V. cholerae O1 strains [29]. Comparison of these restriction enzyme profiling could indicate whether isolates are epidemiologically linked to understanding regional diversity and global distribution for comprehensive ancestry analysis of pathogenic Vibrio spp. [30]. Therefore, this study assesses the polymorphism and mutational diversity of the nucleotide and putative amino acid sequences of virulence (vcgCPI and vcgCPE) and resistance determinants (aac(3)-IIa, (aacC2, strA, Sul 1, and 11) found among human pathogenic Vibrio species that were recovered from surface waters in South-Western districts of Uganda.
Methods
Collection of samples, processing, and enumeration of Vibrio spp.
A total of 230 water samples were collected from 46 villages between June 2018 and October 2018. Using sterilized Nalgene glass bottles, (1000 ml) water samples were collected from different sampling points in each of the four districts (including, Bushenyi, Mitooma, Rubirizi, and Sheema) in South West of Uganda and transported in an ice-cool box to the laboratory for analysis within 6 h. tenfold dilutions were carried out on the water samples as described by Adefisoye and Okoh (2016) [31], 1 mL of each serial dilution was plated onto TCBS agar (thiosulphate citrate bile salts sucrose) (Neogen, Lansing, MI 48912 USA) in triplicates for 24 h and incubated at 37 °C. The presumptive Vibrio spp., was then counted and measured in colony-forming units per milliliters (CFU/mL) of water samples for the yellow and green colonies identified by colonial morphology and cultural characteristics of the colony as described by Pfeffer and Oliver (2003) and Kriem et al., (2015) [32, 33]. A single colony of presumptive isolates was then subcultured onto nutrient agar to ascertain purity, and each pure culture was picked and stored in glycerol stock for further analysis.
Molecular confirmation of presumptive Vibrio species
The glycerol stocks were resuscitated using nutrient broth (Merck, Modderfontein, South Africa) and incubated for 24 h at 37 °C, while the genomic DNA of the 981 presumptive Vibrio spp., isolates were extracted following the boiling procedure described by [2, 34] with slight modifications. The fresh overnight bacterial isolates were sub-cultured into sterile 1.5 mL microfuge tubes and centrifuged (HERMLE, Siemensstr-25, D-78564 Wehingen, Germany) at a speed of 13,000 rpm for 10 min. The cell pellets were washed twice with phosphate-buffered saline, resuspended in 500 µL sterile distilled water, and then lysed to release the DNA by boiling at 100 °C for 10 min in pre-heated heating blocks (Techne heating block Dri-Block, DB-3D; Gauteng, Pretoria, South Africa). Afterward, the suspensions were centrifuged for 5 min at 15,000 rpm, and the supernatant was carefully pipetted into sterile Cryon tubes (Labotec, South Africa) and stored at − 20 °C.
The primer pair F-5′CGG TGA AAT GCG TAG AGA T-3′ and R-5′TTA CTA GCG ATT CCG AGT TC-3′ previously described by [35], was purchased from Inqaba Biotechnical Industries (Pty) Ltd., Pretoria, South Africa and used to amplify 16S rRNA genes of Vibrio spp., generating an amplicon size of ~ 663. The PCR reaction mixture of 25 µL (12 µL PCR master mix (New England BIOLABS), 1 µL of each forward and reverse primers, 6 µL of PCR grade water, and 5 µL of genomic DNA template were amplified using BioRad T100 thermal Cycler Lasec. (621BR44012, Singapore). Afterwards, 4 µL of the amplicons were electrophoresed in 1.5% agarose gel using the thermal tank (Labnet, Enduro Gel XL, USA) on staining with ethidium bromide (0.5 µL) and 0.5X Tris–borate EDTA (TBE) buffer with a controlled base size of 100-bp DNA ladder (New England BIOLABS), Madison, WI, USA). A 100 V and 60 min electrophoresis process was done, and the gels were visualized under the UV trans-illuminator (Alliance 4.7, UVItec, Merton, London, UK.
Determination of virulence genes signature of the confirmed Vibrio species
The virulence gene signature distributions in the confirmed Vibrio spp isolates were determined using the PCR technique as we have described before [2, 36], with slight modifications. The set of primers indicates the targeted genes, sequence, and conditions in Table 1. The PCR reaction mixture was made up to a final volume of 25 μL, while the electrophoresed amplified amplicons were visualized as stated earlier.
Antibiotic resistance determinants using simplex PCR
The simplex PCR was used to assay relevant resistance determinants for the isolates obtained from phenotypic antibiotic-resistant Vibrio spp., isolates based on the susceptibility patterns [9, 34]. The resistance genes for the classes and specific antibiotics were assayed for including those of aminoglycosides [Kanamycin, Nitrofurantoin (strA, aadA, aac(3)-IIa (aacC2)a)]; and sulfonamides [Trimethoprime-sulfamethoxazole (sul11)]. The primers targeting conserved regions of the specific genes, sequence, cycle procedures, and expected amplicon band sizes are indicated in Table 1. All the PCR and electrophoresis procedures were carried out as earlier described.
Partial nucleotide sequencing of amplicons and sequence analysis
For sequencing of amplicon gene analyses, the positive PCR products/amplicons of high quality were selected for sequencing at Inqaba Biotechnical Industries (Pty) Ltd. (Hatfield 0028, South Africa) using the forward and reverse primers earlier used in PCR amplification [40]. The amplicons/PCR products were purified and sequenced with standard Sanger sequencing [41]. Sequenced DNA were cleaned and edited in Bio Edith 3.3.19.0 and chromas 2.6.6 software, then blasted and assembled using Geneious 2021.1 [42]. As a first step, the DNA sequences were run via the Basic Local Alignment Search Tool (BLAST) to ensure that all of the sequences were genuinely Vibrio spp., compared to other GenBank sequences. Bioedit software [43] was used for nucleotide sequence alignment, whereas ClustalW, implemented in Geneious 10.1.2 software, was used for amino acid alignment [42].
Restriction enzymes length polymorphism (RFLP) using six different digestive enzymes
The consensus sequence generated from Bioedit was used to analyze for RFLP by exploring the New England Biolabs restriction enzymes tools for analyzing DNA sequences at the site: http://nc2.neb.com/NEBcutter2/. 6 custom digest restriction enzymes were used to cut the DNA sequences, and predict the respective enzymes’ gel banding patterns [44]. The number of banding patterns produced per sequence was then counted and recorded respectively.
Results
A. Multiple alignments of the V. vulnificus virulence gene and three different isolates of the resistance genes
The gene investigated includes; [(aac(3)-IIa, (aacC2)a] aminoglycoside N(3)-acetyltransferase III, [strA] aminoglycoside 3′-phosphotransferase and [aadA] aminoglycoside (3′′) (9) adenylyltransferase, both resistance genes of (kanamycin, nitrofurantoin) aminoglycosides, and [sul 1 and 11] dihydropteroate synthase type 1 and 11 resistance gene of (trimethoprim-sulfamethoxazole) sulfonamides versus NCBI reference bacteria.
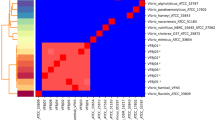
The multiple sequence alignment of the V. vulnificus virulence gene (vcgCPI) represented as (VC) genes obtained from Vibrio isolates in this study and other reference bacterial species show numerous nucleotide variations at different locations (Fig. 1). However, the nucleotide sequence polymorphism and mutation only result in similar putative amino acids in the virulence reference isolates, such as K. grimontii (LR607341) and K. huaxiensis (CP036175) at nucleotide positions 73–75 (A → G) and 300–302 (N → S) (Figs. 1 and 2).
Putative amino acid sequences of the aligned V. vulnificus virulence gene (vcgCPI) as obtained in Geneious [42]
Figure 3 shows the nucleotide and amino acid sequence alignment of the five aminoglycosides resistance gene (strA) of Vibrio spp represented as (SR) obtained from Vibrio isolates in this study and seven other reference bacterial species. It could be deduced that the partial SR gene region of the Vibrio isolates sequenced is highly conserved; no single nucleotide difference was observed among the five sequences compared with all the reference bacterial species analyzed. Also, the putative amino acid sequences of the aligned SR are shown in the Supplementary file.
Nucleotide sequence alignment of partial five aminoglycosides resistance gene (strA) of Vibrio spp (SR) genes obtained from Vibrio isolates with sequences of different reference bacterial species from the GenBank.
The outcome of the nucleotide sequence alignment of the aminoglycosides resistance gene (aadA) of Vibrio spp., represented as (a) gene, from the Vibrio isolates in this study, with other six different reference bacteria species from the GenBank, equally showed high-level genome conservation across the different bacterial species, as only one nucleotide difference was observed at position 482 (A → G) for both a463 and Aeromonas salmonicida (AF327727) (Fig. 4). However, the nucleotide difference does not vary in the amino acid sequence at the different bacterial species (Fig. 5).
The nucleotide sequence alignment of the 11 aminoglycosides resistance gene (sul 1 and 11) represented as (S) genes obtained from Vibrio isolates in this study and five different reference bacterial isolates from the GenBank, equally showed a high level of conservancy with only two observed regions of nucleotide polymorphism (102 and 140) as shown in Fig. 6. Sequence S414 from this study has nucleotide ‘C’ at position 102 alongside the reference C. freundii (KY986974), while other reference bacterial species and the remaining ten sequences obtained in this study have ‘T’ at the same position. Also, at position 140, the sequence S414 has ‘A’ together with the reference C. freundii (KY986974) and P. mirabilis (MT585156), while other sequences have nucleotide ‘C’ at the same position (Fig. 6). However, amino acid differences only exist due to the nucleotide polymorphism at position 140 (A → E), as shown in Fig. 7. The MSA was done in Geneious Prime 2021.0.3 [42].
B. Restriction enzymes length polymorphism using six different digestive enzymes
The result of the banding patterns as produced by the restriction enzymes show no significant differences among the Vibrio isolates and reference bacterial isolates extracted from the GenBank. While only the banding pattern produced in isolate a3_966, when digested by Hinf1, was significantly different with one band compared to the others and referenced bacterial isolates from the GenBank and when the restriction enzymes were combined as seen in A (MT151380 V. cholerae) (Table 2).
The banding patterns produced by the restriction enzyme endonuclease digestion of virulence-correlated gene (vcgCPI) differed significantly. The isolate VC_181 showed a higher banding pattern to the restriction enzymes, i.e., HinP1I, MwoI, and StyD4I, compared to others used and the reference bacterial isolates from the GenBank. In comparison, the banding patterns produced no significant differences when the isolates were digested with the enzymes DpnI and Hinf1. However, the reference bacteria CP071393 K. Michigan; CP036175_K. huaxiensis produced only a single band when digested with the StyD4I restriction enzyme in Table 3.
The banding patterns produced by the restriction enzyme digestion of protein dihydropteroate synthase type 1 and type II genes (Sul 1 and 11) differed significantly. The isolate S_406 showed no band when digested with the restriction enzyme NlaIV compared to others and the reference bacterial isolates from the GenBank. Generally, variably different banding patterns were observed among all the isolates when digested with the enzymes DpnI, HinP1I, NlaIV, MwoI, and StyD4I, as shown in Table 4.
Discussion
Since the endemic of Vibrio spp, phenotype variation is frequently used to determine or measure pathogenicity, intraspecies diversity by utilizing metabolizable substrates [45], colony morphotype [46], the presence of membrane proteins and lipopolysaccharide [47], extracellular enzymes such as cytolysins [48, 49], siderophores [50], virulence in mice [51], and resistance to animal host defense systems [52, 53], genetic divergence remain a prompt strategy for virulence determination. The preliminary phenotypic only provides appreciated evidence about the incidence and occurrence of phenotypic identities among V. strains. However, there is still a dearth of information on the characteristics of species mutation in order to predict strain pathogenicity and antibiotic treatment efficacy accurately.
The nucleotide and amino acid alignment results depict a diversity of alterations and mutations in the V. vulnificus virulence (vcgCPI) gene. Among the alterations, only the mutation at codons 309 nucleotide bases significantly affects the protein function of S (serine) compared to others. However, epidemiological studies have implicated the vcgC in clinical Vibrio isolates while the vcgE documented in environment isolates [39, 54]. The nucleotide polymorphisms observed within the genetic loci vcg allele show an incomparable likeness to the genetic characteristics frequently found in environmental isolates, as previously reported by D’souza et al. 2020) [55]. The nucleotides and amino acid alignment of [strA] Aminoglycoside 3′-phosphotransferase show no mutation or alteration in the gene sequences, possibly due to the highly conversed regions of the targeted gene.
The gene [aadA] Aminoglycoside (3′′) (9) adenylyltransferase shows a significant mutation at codon 482, which indicates a change in protein function of (Lys/K) Lysine found in the reference bacteria to (Arg/R) Arginine in the isolate a463. This observation may play a complementary protagonist in advancing high levels of aminoglycosides (e.g., Kanamycin and Nitrofurantoin) resistance, similar to the report shown by Minarini and Darini (2012) quinolone and ciprofloxacin resistance. Similarly, a significant mutation was observed in (sul 1 and 11) Dihydropteroate synthase type 1 and 11 genes at codons 102 and 140 of the isolate S_414. This alteration in codon 102 (T-C) was insignificant, as no implication was found in the putative amino acid. Nevertheless, the alteration at codon 140 (C–A) significantly affects the protein function causing a mutation of (Pro/P) Proline to (His/H) Histidine. This result is similar to the previous findings by Weigel and colleagues, which suggested that a substitution or mutation in an amino acid is sufficient to generate a significant degree of resistance to antibiotics, such as mutations in (Ser-83) for nalidixic acid resistance and in Thr83-Ile resistance to fluoroquinolones [56], alteration in Thr83-Ile resistance to ciprofloxacin [57].
The application of the RFLP technique to determine genomic relatedness of virulence or resistance genes and determine polymorphism among isolated Vibrio spp at various loci have been previously documented, e.g., [58, 59]. The results of restriction enzyme digestions by DpnI, EcoRV, Hinf1, HinP1I, NlaIV, and Sac11 as well as in combinations as utilized in this study revealed that the majority of the Vibrio strains and the reference strains examined share similarity among the selected endonucleases. Specifically, the gene (aac(3)-IIa, (aacC2)a showed a difference of two bands when digested by hint restriction enzymes. This homology may be due to spontaneous translucent isolate, as previously observed by [51, 60]. Most Vibrio strains that have been previously reported are translucent strains, which are different from their opaque parent in the number of capsules produced. Therefore, the results from this study may be very likely due to the differences in physiological characteristics exhibited by these recent isolates. The different banding patterns observed in this study imply that these isolates possess unique pathogenic/biochemical characteristic polymorphism. As the different restriction enzymes (i.e., DpnI, EcoRV, Hinf1, HinP1I, NlaIV, Sac11) tested with nucleotide sequences from three isolates with the same (aac(3)-IIa, (aacC2) an Aminoglycoside N(3)-acetyltransferase III gene, the enzymes (DpnI, HinP1I, NlaIV, MwoI, StyD4I) tested on several isolates protein dihydropteroate synthase type 1 and type II (Sul 1 and 11) genes, and vcgCPI of which the endonuclease(s) produced polymorphic locus of DNA fragments.
Pulsed-field gel electrophoresis (PFGE) has been effectively used to discriminate strains of Vibrio as a powerful tool for differentiating bacterial strains [58, 61,62,63] after genomic cleavage of site-specific, low-frequency restriction endonucleases, exploiting the basic principle of movement of large DNA fragments in gels. This approach is extensively applied in epidemiological studies and less relatively employed in environmental investigation. Therefore, the observed polymorphism among RFLP profiles for tested genes in these environmental Vibrio strains indicates a polymorphic pathogenic/virulence relevance and treatment/management pattern.
Conclusion
In conclusion, the isolates recovered from the surface water in greater Bushenyi encompass a very diverse population of Vibrio spp strains, and those specific subclasses of strains are pathogens that appear to be linked with human disease. The described virulence and resistance determinants possess specific polymorphic locus that may be relevant in pathogenomics, pharmacogenomics, vaccine production, and the control of strains in the future. Ultimately, this approach can provide scientific and rational bases for risk assessment.
Availability of data and materials
The datasets/information used for this study are available from the corresponding author on reasonable request.
References
Janda JM, Powers C, Bryant RG, Abbott SL (1988) Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. https://doi.org/10.1128/CMR.1.3.245
Onohuean H, Okoh AI, Nwodo UU. Epidemiologic potentials and correlational analysis of Vibrio species and virulence toxins from water sources in greater Bushenyi districts, Uganda. Sci Rep 2021:17. https://doi.org/10.1038/s41598-021-01375-3.
Lee MT, Dinh AQ, Nguyen S, Krucke G, Tran TT (2019) Late-onset Vibrio vulnificus septicemia without cirrhosis. Baylor Univ Med Cent Proc. https://doi.org/10.1080/08998280.2019.1580661
Cabanillas-Beltrán H, Llausás-Magaña E, Romero R, Espinoza A, García-Gasca A, Nishibuchi M et al (2006) Outbreak of gastroenteritis caused by the pandemic Vibrio parahaemolyticus O3:K6 in Mexico. FEMS Microbiol Lett. https://doi.org/10.1111/j.1574-6968.2006.00475.x
Velazquez-Roman J, León-Sicairos N, Hernández-Díaz L de J, Canizalez-Roman A. Pandemic Vibrio parahaemolyticus O3: K6 on the American continent. Front Cell Infect Microbiol 2014. https://doi.org/10.3389/fcimb.2013.00110.
Igere BE, Onohuean H, Nwodo UU. Water bodies are potential hub for spatio-allotment of cell-free nucleic acid and pandemic: a pentadecadal (1969–2021) critical review on particulate cell-free DNA reservoirs in water nexus. Bull Natl Res Cent 2022;46. https://doi.org/10.1186/s42269-022-00750-y.
Onohuean H, Agwu E, Nwodo UU (2022) A global perspective of vibrio species and associated diseases: three-decade meta-synthesis of research advancement. Environ Health Insights. https://doi.org/10.1177/11786302221099406
Smirnova NI, Kirillina OA, Kutyrev VV (2000) Genetic markers of epidemic strains of Vibrio cholerae. Zh Mikrobiol Epidemiol Immunobiol (5):87–91.
Onohuean H, Okoh AI, Nwodo UU (2021) Antibiogram signatures of Vibrio species recovered from surface waters in South Western districts of Uganda: Implications for environmental pollution and infection control. Sci Total Environ 807:150706. https://doi.org/10.1016/j.scitotenv.2021.150706
Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T et al (2000) Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. https://doi.org/10.1128/jcm.38.2.578-585.2000
Nasu H, Iida T, Sugahara T, Yamaichi Y, Park KS, Yokoyama K et al (2000) A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol. https://doi.org/10.1128/jcm.38.6.2156-2161.2000
Bier N, Diescher S, Strauch E (2015) Multiplex PCR for detection of virulence markers of Vibrio vulnificus. Lett Appl Microbiol. https://doi.org/10.1111/lam.12394
Guardiola-Avila I, Acedo-Felix E, Sifuentes-Romero I, Yepiz-Plascencia G, Gomez-Gil B, Noriega-Orozco L (2016) Molecular and genomic characterization of vibrio mimicus isolated from a frozen shrimp processing facility in Mexico. PLoS ONE. https://doi.org/10.1371/journal.pone.0144885
Falbo V, Carattoli A, Tosini F, Pezzella C, Dionisi AM, Luzzi I (1999) Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.43.3.693
Davison J (1999) Genetic exchange between bacteria in the environment. Plasmid. https://doi.org/10.1006/plas.1999.1421
Rendueles O, de Sousa JAM, Bernheim A, Touchon M, Rocha EPC (2018) Genetic exchanges are more frequent in bacteria encoding capsules. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007862
Thungapathra M, Amita, Sinha KK, Chaudhuri SR, Garg P, Ramamurthy T, et al. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob Agents Chemother 2002. https://doi.org/10.1128/AAC.46.9.2948-2955.2002.
Iwanaga M, Toma C, Miyazato T, Insisiengmay S, Nakasone N, Ehara M (2004) Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.48.7.2364-2369.2004
Burrus V, Waldor MK (2004) Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. https://doi.org/10.1016/j.resmic.2004.01.012
Amita, Chowdhury SR, Thungapathra M, Ramamurthy T, Nair GB, Ghosh A. Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae 0139 outbreak, Calcutta, India. Emerg Infect Dis 2003. https://doi.org/10.3201/eid0904.020317.
González-Plaza JJ, Šimatović A, Milaković M, Bielen A, Wichmann F, Udiković-Kolić N (2018) Functional repertoire of antibiotic resistance genes in antibiotic manufacturing effluents and receiving freshwater sediments. Front Microbiol. https://doi.org/10.3389/fmicb.2017.02675
Ju F, Beck K, Yin X, Maccagnan A, McArdell CS, Singer HP et al (2019) Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. https://doi.org/10.1038/s41396-018-0277-8
Onohuean H. Occurrence, Antibiotic Susceptibility and Genes Encoding Antibacterial Resistance of Salmonella spp . and Escherichia coli From Milk and Meat Sold in Markets of Bushenyi District , Uganda 2022. https://doi.org/10.1177/11786361221088992.
Onohuean H, Agwu E, Nwodo UU (2022) Systematic review and meta-analysis of environmental Vibrio species – antibiotic resistance. Heliyon 8:e08845. https://doi.org/10.1016/j.heliyon.2022.e08845
Igbinosa EO, Okoh AI (2010) Vibrio fluvialis: an unusual enteric pathogen of increasing public health concern. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph7103628
Ismail H, Smith AM, Sooka A, Keddy KH (2011) Genetic characterization of multidrug-resistant, extended-spectrum-β- lactamase-producing Vibrio cholerae O1 outbreak strains, Mpumalanga, South Africa, 2008. J Clin Microbiol. https://doi.org/10.1128/JCM.00293-11
Korzheva N, Davies TA, Goldschmidt R (2005) Novel Ser79Leu and Ser81Ile substitutions in the quinolone resistance-determining regions of ParC topoisomerase IV and GyrA DNA gyrase subunits from recent fluoroquinolone-resistant Streptococcus pneumoniae clinical isolates. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.49.6.2479-2486.2005
Fukushima K, Saito T, Kohyama A, Watanabe K (2020) Increased quinolone-resistant mutations of gyrA and parC genes after pouchitis treatment with ciprofloxacin. Dig Surg. https://doi.org/10.1159/000504750
Johura FT, Biswas SR, Rashed SM, Islam MT, Islam S, Sultana M et al (2022) Vibrio cholerae O1 El Tor strains linked to global cholera show region-specific patterns by pulsed-field gel electrophoresis. Infect Genet Evol. https://doi.org/10.1016/j.meegid.2022.105363
Rogers L, Power K, Gaora PO, Fanning S (2015) Escherichia coli and Other Enterobacteriaceae: occurrence and detection. Encycl Food Heal. https://doi.org/10.1016/B978-0-12-384947-2.00259-2
Adefisoye MA, Okoh AI (2016) Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiol Open. https://doi.org/10.1002/mbo3.319
Pfeffer C, Oliver JD (2003) A comparison of thiosulphate-citrate-bile salts-sucrose (TCBS) agar and thiosulphate-chloride-iodide (TCI) agar for the isolation of Vibrio species from estuarine environments. Lett Appl Microbiol. https://doi.org/10.1046/j.1472-765X.2003.01280.x
Kriem MR, Banni B, El Bouchtaoui H, Hamama A, El Marrakchi A, Chaouqy N, et al. Prevalence of Vibrio spp. in raw shrimps (Parapenaeus longirostris) and performance of a chromogenic medium for the isolation of Vibrio strains. Lett Appl Microbiol 2015. https://doi.org/10.1111/lam.12455.
Igere BE, Okoh AI, Nwodo UU (2020) Antibiotic susceptibility testing (AST) reports: a basis for environmental/epidemiological surveillance and infection control amongst environmental vibrio cholerae. Int J Environ Res Public Health 17:1–23. https://doi.org/10.3390/ijerph17165685
Igbinosa EO, Okoh AI (2008) Emerging Vibrio species: an unending threat to public health in developing countries. Res Microbiol. https://doi.org/10.1016/j.resmic.2008.07.001
Adefisoye MA, Okoh AI (2017) Ecological and public health implications of the discharge of multidrug-resistant bacteria and physicochemical contaminants from treated wastewater effluents in the Eastern Cape, South Africa. Water (Switzerland). https://doi.org/10.3390/w9080562
Srinivasan V, Gillespie BE, Lewis MJ, Nguyen LT, Headrick SI, Schukken YH et al (2007) Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli isolated from dairy cows with mastitis. Vet Microbiol. https://doi.org/10.1016/j.vetmic.2007.04.040
Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L et al (2004) Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol. https://doi.org/10.1128/JCM.42.12.5444-5452.2004
Rosche TM, Yano Y, Oliver JD (2005) A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol Immunol. https://doi.org/10.1111/j.1348-0421.2005.tb03731.x
Johnning A, Kristiansson E, Fick J, Weijdegård B, Larsson DGJ (2015) Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Front Microbiol. https://doi.org/10.3389/fmicb.2015.01355
Furutani S, Furutani N, Kawai Y, Nakayama A, Nagai H (2022) Rapid DNA sequencing technology based on the sanger method for bacterial identification. Sensors. https://doi.org/10.3390/s22062130
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647. https://doi.org/10.1093/BIOINFORMATICS/BTS199
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp Ser 41:95–98.
Chatterjee S, Raval IH (2019) Pathogenic microbial genetic diversity with reference to health. Microb Divers Genomic Era. https://doi.org/10.1016/B978-0-12-814849-5.00032-0
Kim HJ, Cho JC (2015) Genotypic diversity and population structure of Vibrio vulnificus strains isolated in Taiwan and Korea as determined by multilocus sequence typing. PLoS ONE. https://doi.org/10.1371/journal.pone.0142657
Brown RN, Gulig PA (2009) Roles of RseB, σE, and DegP in virulence and phase variation of colony morphotype of Vibrio vulnificus. Infect Immun. https://doi.org/10.1128/IAI.00205-09
Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae: I. Physical and chemical characterization. Biochim Biophys Acta - Mol Basis Dis 2003. https://doi.org/10.1016/j.bbadis.2003.08.004.
Elluri S, Enow C, Vdovikova S, Rompikuntal PK, Dongre M, Carlsson S, et al. Outer membrane vesicles mediate transport of biologically active Vibrio cholerae cytolysin (VCC) from V. cholerae strains. PLoS One 2014. https://doi.org/10.1371/journal.pone.0106731.
Galvis F, Barja JL, Lemos ML, Balado M (2021) The vibriolysin-like protease VnpA and the collagenase ColA are required for full virulence of the bivalve mollusks pathogen vibrio neptunius. Antibiotics. https://doi.org/10.3390/antibiotics10040391
Thode SK, Rojek E, Kozlowski M, Ahmad R, Haugen P (2018) Distribution of siderophore gene systems on a Vibrionaceae phylogeny: database searches, phylogenetic analyses and evolutionary perspectives. PLoS ONE. https://doi.org/10.1371/journal.pone.0191860
Pettis GS, Mukerji AS (2020) Structure, function, and regulation of the essential virulence factor capsular polysaccharide of vibrio vulnificus. Int J Mol Sci. https://doi.org/10.3390/ijms21093259
Tamplin ML, Jackson JK, Buchrieser C, Murphree RL, Portier KM, Gangar V et al (1996) Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl Environ Microbiol. https://doi.org/10.1128/aem.62.10.3572-3580.1996
Wahlig TA, Stanton E, Godfrey JJ, Stasic AJ, Wong ACL, Kaspar CW (2021) A single nucleotide polymorphism in lptG increases tolerance to bile salts, acid, and staining of calcofluor-binding polysaccharides in Salmonella enterica Serovar Typhimurium E40. Front Microbiol. https://doi.org/10.3389/fmicb.2021.671453
Abdelbary MMH, Feil EJ, Senn L, Petignat C, Prod’hom G, Schrenzel J, et al. Phylogeographical analysis reveals the historic origin, emergence, and evolutionary dynamics of methicillin-resistant Staphylococcus aureus ST228. Front Microbiol 2020. https://doi.org/10.3389/fmicb.2020.02063.
D’souza C, Prithvisagar KS, Deekshit VK, Karunasagar I, Karunasagar I, Kumar BK (2020) Exploring the pathogenic potential of vibrio vulnificus isolated from seafood harvested along the mangaluru coast, india. Microorganisms. https://doi.org/10.3390/microorganisms8070999
Minarini LAR, Darini ALC (2012) Mutations in the quinolone resistance-determining regions of gyrA and parC in enterobacteriaceae isolates from Brazil. Brazilian J Microbiol. https://doi.org/10.1590/S1517-83822012000400010
Weigel LM, Steward CD, Tenover FC (1998) gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.42.10.2661
Prevost G, Jaulhac B, Piemont Y (1992) DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. https://doi.org/10.1128/jcm.30.4.967-973.1992
Tan CW, Rukayadi Y, Hasan H, Abdul-Mutalib NA, Jambari NN, Hara H et al (2021) Isolation and Characterization of Six Vibrio parahaemolyticus Lytic Bacteriophages From Seafood Samples. Front Microbiol. https://doi.org/10.3389/fmicb.2021.616548
Mohamad N, Amal MNA, Saad MZ, Yasin ISM, Zulkiply NA, Mustafa M, et al. Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet Res 2019. https://doi.org/10.1186/s12917-019-1907-8.
Oliver JD, Jones JL (2014) Vibrio parahaemolyticus and Vibrio vulnificus. Mol Med Microbiol. https://doi.org/10.1016/B978-0-12-397169-2.00066-4
Abdelbary MMH, Basset P, Blanc DS, Feil EJ (2017) The evolution and dynamics of methicillin-resistant Staphylococcus aureus. Genet Evol Infect Dis Second Ed. https://doi.org/10.1016/B978-0-12-799942-5.00024-X
Baig S, Rhod Larsen A, Martins Simões P, Laurent F, Johannesen TB, Lilje B et al (2020) Evolution and population dynamics of clonal complex 152 community-associated methicillin-resistant Staphylococcus aureus. MSphere. https://doi.org/10.1128/msphere.00226-20
Acknowledgements
We are grateful to the Infectious Diseases and Medicinal Plants Research Niche Area and Govan Mbeki Research and Development Centre (GMRDC), University of Fort Hare for funding support.
Consent to participate
Not applicable.
Funding
This research was supported by the Infectious Diseases and Medicinal Plants Research Niche Area and the Govan Mbeki Research and Development Centre (GMRDC), University of Fort Hare (UFH).
Author information
Authors and Affiliations
Contributions
Conceptualization, O.H., N.U.U.; methodology, O.H., N.U.U; investigation, O.H.; data curation, O.H.; writing—original draft preparation, O.H.; writing—review and editing, N.U.U.; supervision, N.U.U.; project administration, N.U.U; funding acquisition, N.U.U. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethical committee of the Kampala International University, Western Campus, Uganda, with the clearance number Nr.UG-REC-023/201919.
Consent for publication
All the authors have read and agreed to the final copy of the findings as contained in the manuscript.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Supplementary file 1. The putative amino acid sequences of the aligned SR
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Onohuean, H., Nwodo, U.U. Polymorphism and mutational diversity of virulence (vcgCPI/vcgCPE) and resistance determinants (aac(3)-IIa, (aacC2, strA, Sul 1, and 11) among human pathogenic Vibrio species recovered from surface waters in South-Western districts of Uganda. J Genet Eng Biotechnol 21, 94 (2023). https://doi.org/10.1186/s43141-023-00554-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-023-00554-1