Abstract
Background
Rg3-ginsenoside, a protopanaxadiol saponin, is a well-known adaptogen used for the prevention of cancer and inflammation. However, despite its distinct biological activity, the concentration of Rg3 in the total ginseng extract is insufficient for therapeutic applications. This study aims to convert PPD-class of major ginsenosides into a mixture of minor ginsenoside, to analyze its immune-regulatory role in macrophage cells.
Results
Using heat and organic acid treatment, three major ginsenosides, Rc, Rd, and Rb1, were converted into a mixture of minor ginsenosides, GRg3-mix [Rg3(S), Rg3(R), Rg5, and Rk1]. Purity and content analysis of the transformed compound were performed using thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC), compared with their standards. Preceding with the anti-inflammatory activity of GRg3-mix, lipopolysaccharide (LPS)-stimulated murine RAW264.7 macrophage cells were treated with various concentrations of GRg3-mix (6.25, 12.5, 25, and 50 μg/mL). The cell viability assay revealed that the level of cell proliferation was increased, while the nitric oxide (NO) assay showed that NO production decreased dose-dependently in activated RAW264.7 cells. The obtained results were compared to those of pure Rg3(S) ≥ 98% (6.25, 12.5, and 25 μg/mL). Preliminary analysis of the CCK-8 and NO assay demonstrated that GRg3-mix can be used as an anti-inflammatory mediator, but mRNA and protein expression levels were evaluated for further confirmation. The doses of GRg3-mix significantly suppressed the initially upregulated mRNA and protein expression of inflammation-related enzymes and cytokines, namely inducible nitric oxide synthase (iNOS), cyclooxygenase-2 (COX-2), nuclear transcription factor kappa B (NF-κB), tumor necrosis factor (TNF-α), and interleukins (IL-6 and IL1B), as measured by reverse transcription-polymerase chain reaction and western blotting.
Conclusions
Our pilot data confirmed that the mixture of minor ginsenosides, namely GRg3-mix, has high anti-inflammatory activity and has an easy production procedure.
Similar content being viewed by others
Background
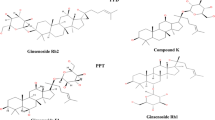
Panax ginseng (family Araliaceae), also known as the “king of herbs,” is a well-known natural and traditional medicine in eastern Asia [1]. Its root consists of organic (80–90%) and inorganic (10%) substances, along with several active saponins, nitrogenous substances, carbohydrates, peptides, amino acids, vitamins, essential oils, and minerals [2]. Among them, ginsenosides are the principal bioactive components and are triterpene saponins that are used as marker compounds for ginseng species [3]. Ginsenosides are categorized into three broad classes based on their hydroxyl group position: protopanaxadiols (PPD), protopanaxatriols, and oleananes [4], as shown in Fig. 1. To date, nearly 200 major and minor ginsenosides have been reported, all of which have a steroidal structure [5]. Because of this structure, they can easily interact with cell membranes, intracellular and extracellular receptors, and membrane-bound ion channels, leading to alterations at the transcriptional level [6]. Based on the percentage of the total ginseng extract, ginsenosides can be divided into two categories: major ginsenosides and minor ginsenosides. Major ginsenosides account for 80% of the total ginseng extract, including Rc, Rd, Rb2, Rb1, Rg1, and Re, whereas minor ginsenosides are present in the trace amounts ≤ 0.2%, including 20(S) and 20(R)- Rg3, Rg5, Rh2, Rk1, Rh1, and Rg2 [7,8,9].
The interest of researchers in the medicinal use of ginseng is increasing owing to its pharmacological effects, which has led to the publication of various studies on its principal active components and their efficacy [10]. The amount of rare minor ginsenosides in the total ginseng extract is insufficient to meet clinical and industrial demands [11]. Therefore, minor ginsenosides (such as F2, Rg3(S), Rg3(R), Rg5, Rk1, and CK) are derived from the major ginsenosides using conversion processes such as biotransformation, chemical or physical conversion, and heat acid treatment [12, 13]. Moreover, studies have demonstrated that absorption of major ginsenosides by the gastrointestinal tract is difficult and the gut microbiome deglycosylases major ginsenosides and ultimately converts them into minor ginsenosides [14, 15].
Prior studies have demonstrated the regulatory role of minor ginsenosides in inflammation [16], cancer [17], obesity [18], diabetes [19], immune stimulation [20], vasodilation [21], and neuroprotection [22], indicating their therapeutic potential. Among the known minor ginsenosides, Rg3 is particularly effective in blocking inflammatory responses [23, 24]. In one study, Rg3 inhibited Th2 cytokine and eotaxin production to repress inflammation and oxidative stress by lowering AHR, eosinophil infiltration, and mucus hypersecretion [25]. The protective and therapeutic potential of Rg3 was demonstrated by the inhibition of oxidants in APAP-induced hepatic damage by alleviating apoptosis and necrosis [26]. The anti-scarring property of Rg3 was demonstrated by the modulation of NF-ĸB/I-ĸB signaling to reduce hypertonic scar formation in rabbit ears [27]. The immune-protective effects of Rg3 and its molecular mechanism in the PI3K/AKT/mTOR pathway have also been reported [28]. Additionally, Rg3 has been shown to protect in vitro expression of arginase-1 in intoxicated peritoneal macrophages to resolve inflammation [29]. Nonetheless, ginsenoside Rk1 has been reported to inhibit LPS-stimulated NF-κB and Jak2/Stat3 pathways in macrophages [30]. In another study, Rk1 inhibited NF-κB and IκB activation, thereby suppressing inflammation [31]. Regarding the anti-inflammatory effects of ginsenoside Rg5, a study proved that Rg5 treatment suppressed NF-κB, iNOS, and COX-2 in HepG-2 cells against hepatitis [32]. Moreover, the protective effects of Rg5 ginsenoside against kidney injury by inhibition of NLRP3 inflammasome activation in a diabetic mouse confirmed its potency to suppress inflammation [33].
To the best of our knowledge, the influence and efficacy of a mixture of transformed minor ginsenosides processed by heat and organic acid treatment of major ginsenosides have rarely been reported. The present study aimed to provide heat and organic acid treatment to the PPD-class of major ginsenosides (Rc, Rd, and Rb1) and transform them into a mixture of minor ginsenosides, that is, the GRg3-mix (Rg3(S), Rg3(R), Rk1, and Rg5), to evaluate its anti-inflammatory role in LPS-activated RAW 264.7 murine macrophages. This study compares the anti-inflammatory effects of the transformed GRg3-mix with those of the extracted Rg3(S) (≥ 98% pure) through a series of cellular reactions, including cell cytotoxicity assay, nitric oxide assay, and mRNA and protein expression levels. Our results will help to investigate the dose-dependent effects of GRg3-mix on LPS-stimulated inflammatory cytokines in RAW264.7 macrophages and the underlying molecular mechanism of these effects. The findings of this study will augment the application of transformed ginsenosides in the pharmaceutical and functional food industries, thereby replacing the conventional procedures of extracting minor ginsenosides, which are expensive and time-consuming.
Methods
Composition of transformed GRg3-mix
The saponins of Korean ginseng, that is, the PPD-class of major ginsenosides, including Rc, Rd, and Rb1, were purchased from AceEMzyme Co., Ltd., Anseong, South Korea. In the preparation of the GRg3-mix, organic acid and heat treatment were applied to the PPD-class of major ginsenosides to transform it into GRg3-mix, as described previously [34]. A 50-g/L PPD-class of major ginsenosides (Rc, Rd, and Rb1) were dissolved in 2% w/v citric acid with distilled water, followed by heating for 15 min at 121°C. Ginsenoside conversion was confirmed using thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC) analyses. In preparation for a stock solution of GRg3-mix, 0.1% dimethyl sulfoxide (DMSO) was used, and Dulbecco’s modified Eagle’s medium (DMEM) was used for further experimental dilutions.
Identification and purity analysis of transformed GRg3-mix
Silica gel plates 60F254 (Merck, Germany) were used for thin-layer chromatography (TLC) analysis with a solvent chloroform-methanol-water (CHCl3-CH3OH-H2O) in the ratio of 65:35:10. Ginsenoside standards were used as a marker to identify GRg3-mix spots (Rg3(S), Rg3(R), Rk1, and Rg5). For the visualization of spots, 10% (v/v) H2SO4 was sprayed on the TLC plates, after which they were heated for 5 min at 110°C. For high-performance liquid chromatography (HPLC) of GRg3-mix, an HPLC system (Younglin Co. Ltd, Korea) with a quaternary pump, automatic injector, and single wavelength UV detector (model 730D) was used. Younglin AutoChro 3000 software was used for peak detection and integration. Prodigy ODS (2) C18 column (5 μm, 150 × 4.6 mm i.d.; Phenomenex, USA) with a guard column (Eclipse XDB C18, 5 μm, 12.5 × 4.6 mm i.d.) was used to perform the separation. The mobile phases used were B (water) and C (acetonitrile). The gradient elution started with 68% solvent B and 32% solvent C; the flow rate was kept at 1.0 mL/min. The absorbance was measured at 203 nm with an injection volume of 25μl for 28 min [35].
Preculturing of RAW264.7 macrophages
Murine macrophage cells (RAW264.7) from the Korean Cell Line Bank, Seoul, South Korea, were precultured in DMEM (GibcoTM, South Korea) augmented with 10% heat-incapacitated fetal bovine serum (FBS, GibcoTM, South Korea), 1% Pen/Strep (GibcoTM, South Korea), and 3.7 mg/mL of NaHCO3 in an incubator at 37 °C with 5% CO2 until the cells reached 70% confluency.
Cytotoxicity assessment of GRg3-mix
The cells were seeded in a microtiter plate (1×105 cells/mL) for overnight surface attachment. The following day, they were treated with different doses of GRg3-mix (0, 6.25, 12.5, 25, 50, and 100 μg/mL) and Rg3(S) (0, 6.25, 12.5, and 25 μg/mL), respectively, and incubated for an hour at 37 °C with 5% CO2. The cells were then induced with 10 μg/mL lipopolysaccharide (LPS, L3129 Sigma-Aldrich) and incubated again for 48 h at 37 °C with 5% CO2. For a stock solution of 1 mg/mL LPS, 10 mg lyophilized LPS was dissolved in 10 mL sterile PBS and further aliquoted into the desired working concentration of 10 μg/mL. After 48 h, a CCK-8 reagent (0.001 g/L, Dojindo, Japan) was added to the treated cells. Optical density was measured at 450nm using an ELISA plate reader (Tecan, Switzerland). The proliferation of the treated cells was calculated as the percentage of control cells.
Nitric oxide determination
To determine the NO produced by the cells, a clean bench with 70% confluent cells was seeded at a concentration of 1×105 cells/mL in a microtiter plate for overnight attachment. The next day, GRg3-mix (0, 6.25, 12.5, 25, and 50 μg/mL) or Rg3(S) (0, 6.25, 12.5, and 25 μg/mL) treatment was administered to the attached cells, along with LPS (10 μg/mL) induction for 48 h. After which, 50 μL supernatant was collected from the treated cells and used for NO determination along with 50μL Griess reagent (Sigma-Aldrich, South Korea). An ELISA microplate reader (Tecan, Switzerland) was used to measure the absorbance at 540 nm. NO produced by the treated cells was expressed as a percentage of control cells.
RNA extraction
A density of 1×105 cells/mL was seeded in a microplate with 6 sample wells and incubated overnight at 37°C with 5% CO2. The attached cells were treated with the desired concentrations of GRg3-mix (0, 6.25, 12.5, 25, and 50 μg/mL). After an hour, LPS (10 μg/mL) activation of macrophages was performed, and the cells were incubated for a further 48 h. Total RNA was isolated from the treated cells using the RNAiso Plus kit (Takara Shuzo Co., Japan) according to the manufacturer’s instructions.
Reverse transcription-polymerase chain reaction (RT-PCR)
cDNA was synthesized using a Revert Aid first-strand cDNA synthesis kit (Thermo Scientific, USA) with a final concentration of 250 ng/μL of the extracted RNA. The final volume of the resultant mixture was incubated at 65 °C for 5 min, followed by an ice shock for 1 min. The reaction mixture (Thermo Fisher Scientific, South Korea) containing 40 U/μL Riboblock-RNAse-inhibitor, 10 mM dNTPs-mix, 200 U/L transcriptional yeast-RevertAid, 5× reaction buffer with MgCl2 for DNase-I (10×), and 0.2 μg/mL random Hexa-primer was added and incubated for 5 min at 25 °C followed by incubation at 42 °C for 60 min. The termination reaction was performed at 95°C for 5 min in a thermal circulator (Bio-Rad C1000TM Thermal Cycler), and the synthesized cDNA was immediately stored at −20°C.
RT-PCR was performed using a premix (MaximeTM RT-PCR kit, South Korea) according to the manufacturer’s instructions. The oligonucleotide sequences of the primers (Cell Signaling Technology, USA) are shown in Table 1 and were used to quantify the following genes: NF-κB, cyclooxygenase-2, inducible nitric oxide synthase (iNOS), tumor necrosis factor-alpha (TNF-α), interleukin-1 beta (IL1 β), interleukin 6 (IL-6), and beta-actin (β-actin) [36]. An initial denaturation of 2 min at 95°C was followed by the following cycle parameters: 1-min annealing of NF-κB (57°C), COX-2 (54°C), iNOS (65°C), TNF-α (57°C), IL-1β (60°C), IL-6 (55°C), and β-actin (56°C) was tailed by 1 min of elongation at 72°C and 1 min of denaturation at 94°C. A 2% agarose gel was used for the separation of the resulting PCR products, while ethidium bromide stained the bands for visualization. All reactions were performed in triplicate. The results were analyzed by comparing the quantification cycle (Cq) value of the genes to that of β-actin.
Western blotting
The cells were seeded at a concentration of 1×105 cells/mL in a six-well microplate and incubated overnight with 5% CO2 at 37°C. The next day, the attached cells were treated with different concentrations of GRg3-mix (0, 6.25, 12.5, 25, and 50 μg/mL) and induced with LPS (10 μg/mL). After 48 h, 200 μL of protein extraction solution (PRO-PREPTM 17081, intron-biotechnology, South Korea) was used to extract protein from the cells, followed by a modified Bradford assay to determine the protein concentration. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the protein (30 μg each), after which samples were transferred to a nitrocellulose membrane (Tech & Innovation). The nitrocellulose membranes were blocked with 5% skim milk, followed by hybridization with the following primary antibodies procured from AbcamTM, South Korea: rabbit-polyclonal anti-NF-κB (ab16502), mouse-monoclonal anti-β actin (ab8226), rabbit-polyclonal anti-COX2 (ab15191), rabbit-polyclonal anti-iNOS (ab3523), mouse-monoclonal anti-IL-6 (ab9324), and mouse-monoclonal anti-TNFα (ab1793). After incubation with horseradish-peroxidase-conjugated secondary antibodies (AB-Frontier-LFQC0103, South Korea), an enhanced chemiluminescence reagent (Westsave Gold, AbFrontier, South Korea) was used to soak the membranes for 5 min, followed by exposure to the radiographic film (Agfa-HealthCare). Protein expression levels were normalized to those of a β-actin control.
Statistical analysis
All experiments were conducted in triplicates, and the results were expressed as the mean ± standard deviation. Duncan’s multiple range test and one-way analysis of variance were performed to evaluate the differences between the control group and GRg3-mix- treated cells. Statistical software (SPSS 19.0, SPSS Inc., Chicago, IllinoisL, USA) pointed out statistically significant values, with p < 0.05.
Results
Composition and identification of GRg3-mix
The PPD-class of major ginsenosides (Rc, Rd, and Rb1) was efficiently transformed into GRg3-mix (Rg3(S), Rg3(R), Rg5, and Rk1) by heat and organic acid treatment, as shown in Fig. 2. TLC analysis indicated two spots of GRg3-mix when the results were compared with the ginsenoside standard. The upper spot contained Rg5+Rk1, while the lower spot contained Rg3(S) and Rg3(R), as shown in Fig. 2A. TLC results were further confirmed by HPLC peaks marked in comparison with the HPLC standard, as shown in Fig. 2B. The transformation pathway of the major PPD-class of major ginsenosides into the minor ginsenoside Rg3-mix after the heat and organic acid treatment is depicted in Fig. 2C.
Conversion of PPD-mix ginsenosides into Rg3-mix ginsenosides. A TLC and B HPLC analysis of heat acid-transformed GRg3-mix, for purity and content check by using ginsenoside standards to compare spots and peaks obtained. C Transformation pathway of major PPD-class of major ginsenosides into minor ginsenoside Rg3-mix [Rg3(S), Rg3(R), Rg5 and Rk1]. std, ginsenoside standard; C, control; S, sample
Effect of GRg3-mix and Rg3(S) on RAW 264.7 cell proliferation
To determine the cytotoxicity of GRg3-mix and the proliferation of RAW 264.7, macrophages were assessed using a CCK-8 reagent, and the results were compared with those of Rg3(S). The results showed that cell viability increased gradually with an increase in the concentration of GRg3-mix (Fig. 3A). At concentrations ranging from 6.25 to 50 μg/mL, cell viability increased by approximately 30–50%, after which a decline in cell proliferation was observed. These results suggest that GRg3-mix has a dose-dependent proliferative effect on murine RAW 264.7 macrophage cells when compared to the control group (naïve) (p < 0.05). On the other hand, cell viability increased in Rg3(S)-treated cells up to 6.25 μg/mL; however, higher concentrations of Rg3(S), i.e., 12.5–25 μg/mL showed decreased cell proliferation (Fig. 3B), indicating that pure Rg3(S) had the highest activity at lower concentrations. However, the cell viability assay results showed that the GRg3-mix has promising anti-inflammatory activity up to 50 μg/mL.
Effect of GRg3-mix and Rg3(S) on cell viability of LPS-activated RAW264.7 macrophages. LPS-induced RAW264.7 macrophages were incubated with different concentrations of GRg3-mix (A) and Rg3(S) (B), to determine cell viability using CCK-8 assay. All values are presented as mean ± SD (n=3). Different superscripts on the bars point out significant differences (p < 0.05). LPS, lipopolysaccharide; SD, standard deviation
Effect of GRg3-mix and Rg3(S) on NO production by RAW 264.7 cells
A Griess reagent kit was used to measure NO production in LPS-induced RAW264.7 cells. A significant increase in NO production was observed upon stimulation with LPS, but GRg3-mix treatment (6.25–50 μg/mL) decreased the production of NO dose-dependently (Fig. 4A). Decreased levels of nitric oxide in LPS-induced RAW 264.7 macrophages to support the fact that GRg3-mix plays a role in suppressing nitric oxide-induced inflammation. Similarly, Rg3(S) treatment (6.25–25 μg/mL) decreased NO production dose-dependently (Fig. 4B), proving its anti-inflammatory role. However, the effect of Rg3(S) on NO production was steady and less significant than that of the GRg3-mix. These results further support our hypothesis that GRg3-mix is an invasive compound to regulate inflammation.
Effect of GRg3-mix and Rg3(S) on NO production in LPS-stimulated RAW264.7 macrophages. The LPS-induced RAW264.7 macrophages were incubated with different concentrations of GRg3-mix (A) and Rg3(S) (B) to determine the level of NO production using the Griess reagent. All values are presented as mean ±SD (n=3). Different superscripts on the bars point out significant differences (p < 0.05). LPS, lipopolysaccharide; NO, nitric oxide; SD, standard deviation
Effect of GRg3-mix on mRNA expression of inflammatory cytokines
The results from the preliminary analysis of CCK-8 and NO production assays were further confirmed by evaluating the mRNA expression of the marker inflammatory cytokines. As LPS induced the mRNA expression of iNOS, NF-κB, IL-6, TNF-α, COX-2, and IL-1β, RT-PCR was performed to visualize their expression. LPS stimulation substantially upregulated gene expression, which was dose-dependently downregulated by treatment with GRg3-mix (6.25–50 μg/mL) (Fig. 5A, B). The results proved that GRg3-mix suppressed LPS-induced inflammatory cytokines by transcriptionally inhibiting their expression.
Effects of GRg3-mix on mRNA expression of LPS-induced RAW 264.7 cells. RAW264.7 macrophages were treated with various concentrations of GRg3-mix (6.25, 12.5, 25, and 50μg/mL) followed by LPS induction. Total RNA was extracted, using an RNAiso Plus kit, after 48 h. RT-PCR was performed to visualize the mRNA expression of inflammatory cytokines (A). Bands were visualized using ImageJ software (B). All values are presented as mean ± SD (n=3). RT-PCR, real-time polymerase chain reaction; LPS, lipopolysaccharide; SD, standard deviation
Effect of GRg3-mix on protein expression of inflammatory cytokines
Protein expression in LPS-induced RAW264.7, treated with GRg3-mix, was found to correlate with mRNA expression. All protein-expressing inflammatory cytokines (iNOS, NF-κB, IL-6, TNF-α, and COX-2) were downregulated upon GRg3-mix (6.25–50 μg/mL) treatment for 48 h. Initially, LPS stimulation upregulated the expression of inflammatory mediators. However, it was gradually downregulated with different concentrations of GRg3-mix (Fig. 6A, B).
Effects of GRg3-mix on protein expression of LPS-induced RAW 264.7 macrophages. RAW264.7 cells were treated with GRg3-mix (6.25, 12.5, 25, and 50μg/mL) followed by LPS activation. After 48 h of incubation, the protein was extracted and quantified using the Bradford assay. SDS-PAGE was performed to visualize the protein expression of inflammatory cytokines (A). Bands were visualized using ImageJ software (B). All values are presented as mean ± SD (n=3). LPS, lipopolysaccharide; PES, protein extraction solution; SD, standard deviation
Discussion
Ginseng is considered a panacea on the Korean Peninsula and consists of various advantageous compounds [37]. Ginsenosides are used to cure all bodily ailments. Scientific advancements have enabled us to explore the biological effects and molecular principles of major or derived minor ginsenosides [38]. Many studies have summarized the efficacy and usefulness of ginseng extract as a whole or as a single extracted ginsenoside [39, 40]. However, there is no study available to prove the in vitro anti-inflammatory effect of ginsenoside mixture (transformed by heat and organic acid treatment); therefore, we performed this study to understand the effect of GRg3-mix in murine RAW 264.7 macrophage cells.
Inflammation is a non-specific immune reaction in response to bodily grievance [41]. LPS can cause inflammation and activate macrophages via pathogen-associated molecular patterns (PAMPs). Hence, macrophages recognize PAMPs by Toll-like receptors, stop proliferation, and activate the biosynthesis of inflammatory mediators, such as interleukins (ILs), TNF-α, and NF-κB pathway, which generates co-stimulatory molecules required for the adaptive immune response against foreign invasion [42]. Owing to their importance in immune scrutiny, macrophages are of great importance for the study of the NF-κB pathway [43].
NO is a gaseous moiety released by macrophages, monocytes, and neutrophils as a defense mechanism against foreign invasion [44]. Prolonged NO emission can stop the normal functioning of the tissue, leading to its degradation [45]. iNOS is an inducible isoform of NOS that catalyzes NO production and is activated in a calcium-independent manner. Excessive NO production is positively correlated with upregulated iNOS expression and vice versa [46]. In this study, we showed that GRg3-mix treatment (6.25–50 μg/mL) significantly downregulated iNOS expression in LPS-induced murine macrophages. In addition, preliminary analysis of the CCK-8 and NO assays also showed that the GRg3-mix has a notable role in mediating the anti-inflammatory response. Hence, this study demonstrated the anti-inflammatory role of GRg3-mix with its key components [Rg3(S), Rg3(R), Rg5, and Rk1].
Prostaglandin E2 (PGE2) is a primary mediator of inflammation, produced by COX-2 [47]. The inducible enzyme “COX-2” is considered the most popular drug target for studying the “commencement and resolution of inflammation,” due to its upregulation in prolonged inflammation [48]. In the current study, the downregulation of mRNA and protein expression of COX-2 suggests that GRg3-mix is a potent compound for the commencement and resolution of inflammation.
NF-κB and its family of transcription factors play a key role in innate immunity and inflammation studies. NF-κB has five proteins in its family, namely, NFκB1 (p50), NFκB2 (p52), Rel-A (p65), Rel-C, and Rel-B. These proteins form homodimer and heterodimer complexes and their activity is controlled by two major pathways: (1) classical/canonical activation and (2) alternative/noncanonical pathways [49]. Macrophages initiate an innate immune response against bacterial pathogens by phagocytosis, thereby eradicating them with the help of reactive oxygen species [50, 51]. TLR-4 is the main mediator of LPS signaling, as it recognizes pathogen-associated molecular patterns, such as bacterial strains, and initiates phosphorylation of interleukin-1 receptor-associated kinases (IRAK-1) [52]. Phosphorylated IRAK-1 is degraded by ubiquitination to activate multimers of the protein complex (TAB1, TAB2, TRAF6, and TAK1). Activated TAK-1 initiates the phosphorylation of IKKs and MKKs [53]. These IKKs play a role in the phosphorylation of IκB-α, which is an inhibitor of NF-κB. In the cytoplasm, the NF-κB p50/p65 complex exists in an inactivated form bound to IκB. Phosphorylation of IκBα leads to ubiquitination and consequent degradation of IκB-α by proteasomes. This degradation sets NF-κB free and translocates it to the nucleus, where it attaches to specific promoter sequences. Activated NF-κB initiates transcription of inflammatory mediators, cytokines, and chemokines (Fig. 7). Hence, increased secretion of ILs and TNF occurs, which provokes an innate immune response [54]. In contrast, activated MKKs phosphorylate and stimulate members of the JNK/p38 MAP kinase (MAPK) family [55].
In the alternate/noncanonical pathway, IKKα is phosphorylated by NF-κB inducing kinase (NIK), which results in the advancement of P100 into mature-P52. Subsequently, NF-κB2 (P52) translocates into the nucleus in the form of a dimer along with RelB, where RelB stimulates promoter activity and P52 pairs up with the specific κB sites of the promoter sequences of the inflammatory cytokines iNOS, COX-2, ILs, and TNFα by stimulating the survival pathway [56]. In this study, the increase in mRNA and protein expression levels of NF-κB in response to LPS stimulation in RAW264.7 macrophage cells was dose-dependently downregulated upon pre-treatment with different concentrations of GRg3-mix. From these findings, we assume that GRg3-mix suppresses inflammatory cytokine expression via the NF-κB-mediated pathway in LPS-activated macrophage cells. In accordance with previous findings [57] and the results of the current study, we deduce that GRg3-mix has a regulatory role on the inflammatory markers and MAPK/NF-κB pathways, either by inhibiting LPS-induced IKK-β phosphorylation or by IRAK activation. However, more research is required to completely understand the mechanism of action of the GRg3-mix and to identify the exact target site of this transformed steroidal compound.
Conclusions
Consequently, research has defined the biological activities of Rg3 ginsenosides, including anti-inflammation, but most studies have reported the efficacy of a single and purified Rg3 compound. The extraction and purification of minor ginsenosides (Rg3) from ginseng is a multistep, time-consuming, and money-consuming process. Using a mixture of major ginsenosides to produce minor ginsenosides with strong anti-inflammatory activity is a promising way to reduce the time, cost, and work procedure. In the present study, the PPD-class of major ginsenosides (Rc, Rd, and Rb1) was converted into a mixture of minor ginsenosides (Rg3(S), Rg3(R), Rg5, and Rk1) by heat and organic acid treatment, which showed a promising ability to combat inflammation. Compared to Rg3(S), GRg3-mix treatment efficiently increased cell viability and decreased NO production in LPS-induced murine RAW264.7 macrophages. In addition, it efficiently downregulated the mRNA and protein expression of the inflammatory cytokines TNF-α, iNOS, NF-κB, IL-6, COX-2, and IL-1β. In conclusion, the economically produced GRg3-mix has promising anti-inflammatory activity with a simple production procedure as compared to pure and extracted Rg3(S) compounds.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Chang-Xiao L, Pei-Gen X (1992) Recent advances on ginseng research in China. J Ethnopharmacol. 36:27–38. https://doi.org/10.1016/0378-8741(92)90057-X
Ratan ZA, Haidere MF, Hong YH, Park SH, Lee JO, Lee J et al (2021) Pharmacological potential of ginseng and its major component ginsenosides. J Ginseng Res. 45:199–210. https://doi.org/10.1016/J.JGR.2020.02.004
Yang W, Qiao X, Li K, Fan J, Bo T, Guo DA et al (2016) Identification and differentiation of Panax ginseng, Panax quinquefolium, and Panax notoginseng by monitoring multiple diagnostic chemical markers. Acta Pharm Sin B. 6:568–575. https://doi.org/10.1016/J.APSB.2016.05.005
Christensen LP (2008) Chapter 1. Ginsenosides: chemistry, biosynthesis, analysis, and potential health effects. Adv Food Nutr Res 55:1–99. https://doi.org/10.1016/S1043-4526(08)00401-4
Mohanan P, Subramaniyam S, Mathiyalagan R, Yang DC (2018) Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J Ginseng Res. 42:123–132. https://doi.org/10.1016/J.JGR.2017.01.008
Han SY, Kim J, Kim E, Kim SH, Seo DB, Kim JH et al (2018) AKT-targeted anti-inflammatory activity of Panax ginseng calyx ethanolic extract. J Ginseng Res. 42:496–503. https://doi.org/10.1016/J.JGR.2017.06.003
Kim JH, Yi YS, Kim MY, Cho JY (2017) Role of ginsenosides, the main active components of Panax ginseng, in inflammatory responses and diseases. J Ginseng Res. 41:435–443. https://doi.org/10.1016/J.JGR.2016.08.004
Kim JH (2018) Pharmacological and medical applications of Panax ginseng and ginsenosides: a review for use in cardiovascular diseases. J Ginseng Res. 42:264–269. https://doi.org/10.1016/J.JGR.2017.10.004
Kim KH, Lee D, Lee HL, Kim CE, Jung K, Kang KS (2018) Beneficial effects of Panax ginseng for the treatment and prevention of neurodegenerative diseases: past findings and future directions. J Ginseng Res. 42:239–247. https://doi.org/10.1016/J.JGR.2017.03.011
Tam DNH, Truong DH, Nguyen TTH, Quynh LN, Tran L, Nguyen HD et al (2018) Ginsenoside Rh1: a systematic review of its pharmacological properties. Planta Med. 84:139–152. https://doi.org/10.1055/s-0043-124087
An KS, Choi YO, Lee SM, Ryu HY, Kang SJ, Yeon Y, Lee YJ, Kang BJ, Choi JE et al (2019) Song KS 2019 Ginsenosides Rg5 and Rk1 enriched cultured wild ginseng root extract bioconversion of Pediococcus pentosaceus HLJG0702: effect on scopolamine-induced memory dysfunction in mice. Nutr 11:1120 11:1120. https://doi.org/10.3390/NU11051120
Zheng MM, Xu FX, Li YJ, Xi XZ, Cui XW, Han CC et al (2017, 2017) Study on transformation of ginsenosides in different methods. BioMed Res Int. https://doi.org/10.1155/2017/8601027
Siddiqi MZ, Ximenes HA, Song BK, Park HY, Lee WH, Han H et al (2021) Enhanced production of ginsenoside Rh2(S) from PPD-type major ginsenosides using BglSk cloned from Saccharibacillus kuerlensis together with two glycosidase in series. Saudi J Biol Sci. 28:4668–4676. https://doi.org/10.1016/J.SJBS.2021.04.079
Tawab MA, Bahr U, Karas M, Wurglics M, Schubert-Zsilavecz M (2003) Degradation of ginsenosides in humans after oral administration. Drug Metab Dispos. 31:1065–1071. https://doi.org/10.1124/DMD.31.8.1065
Cui J (1995) Identification and quantification of ginsenosides in various commercial ginseng preparations. Eur J Pharm Sci. 3:77–85. https://doi.org/10.1016/0928-0987(94)00077-D
Lee IS, Uh I, Kim KS, Kim KH, Park J, Kim Y et al (2016) Anti-inflammatory effects of ginsenoside Rg3 via NF−κB pathway in A549 cells and human asthmatic lung tissue. J Immunol Res. 2016:7521601. https://doi.org/10.1155/2016/7521601
Hong H, Baatar D, Hwang SG (2021) Anticancer activities of ginsenosides, the main active components of ginseng. Evid Based Complement Alternat Med. 2021:8858006. https://doi.org/10.1155/2021/8858006
Park M, Kim KH, Jaiswal V, Choi J, Chun JL, Seo KM et al (2021) Effect of black ginseng and silkworm supplementation on obesity, the transcriptome, and the gut microbiome of diet-induced overweight dogs. Sci Rep. 11:16334. https://doi.org/10.1038/s41598-021-95789-8
Zhao XY, Zhang F, Pan W, Yang YF, Jiang XY (2021) Clinical potentials of ginseng polysaccharide for treating gestational diabetes mellitus. World J Clin Cases. 9:4959–4979. https://doi.org/10.12998/WJCC.V9.I19.4959
Lee YY, Irfan M, Quah Y, Saba E, Kim SD, Park SC et al (2021) The increasing hematopoietic effect of the combined treatment of Korean Red ginseng and Colla corii asini on cyclophosphamide-induced immunosuppression in mice. J Ginseng Res. 45:591–598. https://doi.org/10.1016/J.JGR.2021.02.004
Hyun SH, Bhilare KD, In G, Park CK, Kim JH (2022) Effects of Panax ginseng and ginsenosides on oxidative stress and cardiovascular diseases: pharmacological and therapeutic roles. J Ginseng Res. 46:33–38. https://doi.org/10.1016/J.JGR.2021.07.007
Kim CJ, Ryu HY, Lee S, Lee HJ, Chun YS, Kim JK et al (2021) Neuroprotective effect and antioxidant potency of fermented cultured wild ginseng root extracts of Panax ginseng C.A. Meyer in mice. Molecules. Mol. 26:3001. https://doi.org/10.3390/MOLECULES26103001
Moon KE, Oh HH, Jeong DY, Kim Y (2021) Ginsenoside conversion and anti-inflammatory effect on RAW 264.7 cells of ginseng extract vinegar. J Korean Soc Food Sci Nutr. 50:226–235. https://doi.org/10.3746/JKFN.2021.50.3.226
Ullah HMA, Saba E, Lee YY, Hong SB, Hyun SH, Kwak YS et al (2022) Restorative effects of Rg3-enriched Korean red ginseng and Persicaria tinctoria extract on oxazolone-induced ulcerative colitis in mice. J Ginseng Res. 46:628–635. https://doi.org/10.1016/J.JGR.2021.07.001
Huang WC, Huang TH, Yeh KW, Chen YL, Shen SC, Liou CJ (2021) Ginsenoside Rg3 ameliorates allergic airway inflammation and oxidative stress in mice. J Ginseng Res. 45:654–664. https://doi.org/10.1016/J.JGR.2021.03.002
Gao Y, Yan J, Li J, Li X, Yang S, Chen N et al (2021) Ginsenoside Rg3 ameliorates acetaminophen-induced hepatotoxicity by suppressing inflammation and oxidative stress. J Pharm Pharmacol. 73:322–331. https://doi.org/10.1093/JPP/RGAA069
Ma L, Li LY, Zhao TL (2020) Anti-inflammatory effects of ginsenoside Rg3 on the hypertrophic scar formation via the NF-κB/IκB signaling pathway in rabbit ears. Pharmazie. 75:102–106. https://doi.org/10.1691/PH.2020.9852
Yang J, Li S, Wang L, Du F, Zhou X, Song Q et al (2018) Ginsenoside Rg3 attenuates lipopolysaccharide-induced acute lung injury via MerTK-dependent activation of the PI3K/AKT/mTOR pathway. Front Pharmacol. 9:850. https://doi.org/10.3389/FPHAR.2018.00850
Kang S, Park SJ, Lee AY, Huang J, Chung HY, Im DS (2018) Ginsenoside Rg3 promotes inflammation resolution through M2 macrophage polarization. J Ginseng Res. 42:68–74. https://doi.org/10.1016/J.JGR.2016.12.012
Yu Q, Zeng KW, MA, Ma X, Jiang Y, Tu P, Wang X. (2017) Ginsenoside Rk1 suppresses pro-inflammatory responses in lipopolysaccharide-stimulated RAW264.7 cells by inhibiting the Jak2/Stat3 pathway. Chin. J Nat Med. 15:751–757. https://doi.org/10.1016/S1875-5364(17)30106-1
Li Z, Zhao L, Chen J, Liu C, Li S, Hua M et al (2020) Ginsenoside Rk1 alleviates LPS-induced depression-like behavior in mice by promoting BDNF and suppressing the neuroinflammatory response. Biochem Biophys Res Commun. 530:658–664. https://doi.org/10.1016/J.BBRC.2020.07.098
Lee SM (2014) Anti-inflammatory Effects of ginsenosides Rg5, Rz1, and Rk1: inhibition of TNF-α-induced NF-κB, COX-2, and iNOS transcriptional expression. Phytother Res. 28:1893–1896. https://doi.org/10.1002/PTR.5203
Zhu Y, Zhu C, Yang H, Deng J, Fan D (2020) Protective effect of ginsenoside Rg5 against kidney injury via inhibition of NLRP3 inflammasome activation and the MAPK signaling pathway in high-fat diet/streptozotocin-induced diabetic mice. Pharmacol Res. 155:104746. https://doi.org/10.1016/J.PHRS.2020.104746
Siddiqi MZ, Cui CH, Park SK, Han NS, Kim SC, Im WT (2017) Comparative analysis of the expression level of recombinant ginsenoside-transforming β-glucosidase in GRAS hosts and mass production of the ginsenoside Rh2-Mix. PLoS One. 12:e0176098. https://doi.org/10.1371/JOURNAL.PONE.0176098
Siddiqi MZ, Park HY, Kim G, Cui C, Jo YJ, Kim S et al (2021) Production of the minor ginsenoside F2 from the PPD-mix-type major ginsenosides using a novel recombinant glycoside hydrolase from Novosphingobium aromaticivorans. Biotechnol Bioproc E. 26:956–967. https://doi.org/10.1007/S12257-020-0215-2
Baatar D, Siddiqi MZ, Im WT, Ul Khaliq N, Hwang SG (2018) Anti-inflammatory effect of ginsenoside Rh2-mix on lipopolysaccharide-stimulated RAW 264.7 murine macrophage cells. J Med Food. 21:951–960. https://doi.org/10.1089/JMF.2018.4180
Saba E, Lee YY, Rhee MH, Kim SD (2020) Alleviation of ulcerative colitis potentially through th1/th2 cytokine balance by a mixture of Rg3-enriched Korean red ginseng extract and Persicaria tinctoria. Molecules. Mol. 25:5230. https://doi.org/10.3390/MOLECULES25225230
Yi YS (2019) Ameliorative effects of ginseng and ginsenosides on rheumatic diseases. J Ginseng Res. 43:335–341. https://doi.org/10.1016/J.JGR.2018.04.004
Wee JJ, Park KM, Chung A-S. Biological activities of ginseng and its application to human health. Herb Med BioMol Clin Asp. 2nd ed; 2011, p. 157–74. https://doi.org/10.1201/b10787-9.
Davis MP, Behm B (2019) Ginseng: a qualitative review of benefits for palliative clinicians. Am J Hosp Palliat Care. 36:630–659. https://doi.org/10.1177/1049909118822704
Ferrero-Miliani L, Nielsen OH, Andersen PS, Girardin SE (2007) Chronic inflammation: importance of NOD2 and NALP3 in interleukin-1β generation. Clin Exp Immunol. 147:227–235. https://doi.org/10.1111/J.1365-2249.2006.03261.X
Yang Y, Gu H, Sun Q, Wang J (2018) Effects of Christensenella minuta lipopolysaccharide on RAW 264.7 macrophages activation. Microb Pathog. 125:411–417. https://doi.org/10.1016/J.MICPATH.2018.10.005
Pratidina LA, Wijayanti N, Hwang SG (2021) Anti-inflammatory action of Indonesian black garlic (IBG) ethanol extracts in LPS-stimulated RAW 264.7 macrophage cells. Indonesian J Pharm. 32:201–208. https://doi.org/10.22146/IJP.1658
Saba E, Jeong D, Irfan M, Lee YY, Park SJ, Park CK et al (2018) Anti-inflammatory activity of Rg3-enriched Korean red ginseng extract in murine model of sepsis. Evid Based Complement Alternat Med. 2018:6874692. https://doi.org/10.1155/2018/6874692
Sharma JN, Al-Omran A, Parvathy SS (2008) Role of nitric oxide in inflammatory diseases. Inflammopharmacology. 2007(15):252–259. https://doi.org/10.1007/S10787-007-0013-X
Aktan F (2004) iNOS-mediated nitric oxide production and its regulation. Life Sci. 75:639–653. https://doi.org/10.1016/J.LFS.2003.10.042
Greenhough A, Smartt HJM, Moore AE, Roberts HR, Williams AC, Paraskeva C et al (2009) The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 30:377–386. https://doi.org/10.1093/CARCIN/BGP014
Rumzhum NN, Ammit AJ (2016) Cyclooxygenase 2: its regulation, role and impact in airway inflammation. Clin Exp Allergy. 46:397–410. https://doi.org/10.1111/CEA.12697
Zhou Y, Cui C, Ma X, Luo W, Zheng SG, Qiu W (2020) Nuclear factor κB (NF-κB)–mediated inflammation in multiple sclerosis. Front Immunol. 11:391. https://doi.org/10.3389/FIMMU.2020.00391
Nathan CF, Hibbs JB (1991) Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr Opin Immunol. 3:65–70. https://doi.org/10.1016/0952-7915(91)90079-G
Miller RA, Britigan BE (1997) Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 10:1–18. https://doi.org/10.1128/CMR.10.1.1
Paik YH, Schwabe RF, Bataller R, Russo MP, Jobin C, Brenner DA (2003) Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology. 37:1043–1055. https://doi.org/10.1053/JHEP.2003.50182
Holst B (2013) Study of the capacity of toll-like receptors to modulate pro-inflammatory responses mediated by receptors for the complement anaphylatoxin C5a
Liu C, Tang X, Zhang W, Li G, Chen Y, Guo A et al (2019) 6-Bromoindirubin-3′-oxime suppresses LPS-induced inflammation via inhibition of the TLR4/NF-κB and TLR4/MAPK signaling pathways. Inflammation. 42:2192–2204. https://doi.org/10.1007/S10753-019-01083-1
Lawan A, Bennett AM (2017) Mitogen-activated protein kinase regulation in hepatic metabolism. Trends Endocrinol Metab. 28:868–878. https://doi.org/10.1016/J.TEM.2017.10.007
Tait SWG, Reid EB, Greaves DR, Wileman TE, Powell PP. Mechanism of inactivation of NF-κB by a viral homologue of IκBα: signal-induced release of IκBα results in binding of the viral homologue to NF-κB. J Biol Chem. 2000;275:34656–34664. https://doi.org/10.1074/jbc. M000320200.
Klinke DJ, Ustyugova IV, Brundage KM, Barnett JB (2008) Modulating temporal control of NF-κB activation: Implications for therapeutic and assay selection. Biophys J. 94:4249–4259. https://doi.org/10.1529/BIOPHYSJ.107.120451
Acknowledgements
Not applicable
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the High Value-added Food Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321038-5), and by the Brain-pool (BP) Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (project no. 2019H1D3A1A02070958).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: MZS, WTI, and SGH. Performed the experiments: ZM and JHL. Analyzed the data: MZS, ZM, WTI, and SGH. Wrote the paper: ZM. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marium, Z., Siddiqi, M.Z., Lee, JH. et al. Repressing effect of transformed ginsenoside Rg3-mix against LPS-induced inflammation in RAW264.7 macrophage cells. J Genet Eng Biotechnol 21, 6 (2023). https://doi.org/10.1186/s43141-023-00462-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-023-00462-4