Abstract
Background
Variants in the signal transducer and activator of transcription 4 (STAT4) gene have an important role in the incident of multiple autoimmune diseases including type 1 diabetes mellitus (T1D). It is a genetically related auto-immune disorder that resulted from T cell-mediated destruction of pancreatic cells that are in control for the production of insulin in the blood. The current study aimed to clarify the role of STAT4 (rs7574865) variant allelic and genotypic variations in the susceptibility to type 1 diabetes among Egyptians by using the real-time PCR.
Results
A total of 100 patients and 100 controls were genotyped for rs7574865, and the biochemical and anthropometric parameters were measured to show that type 1 diabetic patients had significantly higher levels of HbA1c and triglycerides compared to non-diabetic individuals (P < 0.05). And genetically, the T allele and GT genotype have a significant correlation with diabetes type 1.
Conclusion
It was confirmed by this study that the rs7574865 T allele and GT genotype have a significant correlation with diabetes type 1 incidence among Egyptian patients.
Similar content being viewed by others
Background
Diabetes is a chronic disease that is characterized by an elevated sugar level in the blood that is either due to compromised insulin excretion from the pancreas or cellular resistance to insulin; it is a multifactorial disease, and generally, it is subdivided into three types (type 1 diabetes, type 2 diabetes, and gestational diabetes) [1]. Diabetes mellitus is considered one of the most important diseases globally because of its high morbidity and major side effects. The number of diabetic patients escalates globally up to 451 million among people between 18 and 99 years old. Moreover, it is expected that when we reach 2045, there will be around 693 million people who will have diabetes mellitus [2]. Type 1 diabetes is an autoimmune disorder developed due to autoimmune destruction of pancreatic beta cells [3];, also, it is triggered due to some other factors in genetically susceptible individuals [1, 4]. Type 1 diabetes mellitus (T1D) is not a widely spread kind of diabetes like type 2, as it represents from 5 to 10% of diabetes mellitus patients [5]. Type 1 diabetes also is the most communal metabolic and endocrine disorder in children and adolescents; its incidence is mostly due to genetic disorders. Genome-wide association studies and meta-analyses identified multiple genetic risk factors for T1D [6]. The main genes causing type 1 diabetes mellitus are found mainly within the major histocompatibility complex region (HLA) [7]. Genetics have a crucial role in the susceptibility of diabetes type 1; the most common genes related to type 1 diabetes are CTLA-4, PTPN22, STAT4, STAT3, and IFIH1. Certain genes are commonly found in one group of people than in another, and that is why race and ethnicity affect disease incidence [8]. Signal transducers and activators of transcription (STAT) proteins are activated by many cytokines, the mechanism of STAT4-mediated IL-12 signaling [9,10,11], and there is a strong evidence that supports a role of IL-12 in autoimmune diabetes [12,13,14]. The signal transducer and activator of transcription 4 (STAT4) gene is in charge of signal transduction of multiple proinflammatory cytokines [15]; its genetic position is 2q32.2-32.3. STAT4 gene is vital in the development of Th1 cells so it plays an important role in immunity, and that is why it was associated with different immune diseases. This study, therefore, inspects the impact of STAT4 variants on the vulnerability of type 1 diabetes among Egyptians.
Methods
Subjects
We recruited 200 un-related Egyptians, 100 T1DM patients who were assured diabetic patients as per the American Diabetes Association (ADA), and 100 healthy individuals. All patients were diagnosed as type 1 diabetic before the age of 15 years and were dependent on exogenously administered insulin. And control was healthy people with no chronic disease such as any other types of diabetes, cardiovascular, renal, and hepatic diseases. No clinical evidence or family history of autoimmune diseases were eligible for inclusion, while non-Egyptians, Egyptian patients with any type of diabetes, and patients with clinical evidence or family history of autoimmune diseases, renal, and hepatic disease were excluded. This research was accepted by the Ethical Committee of the Egyptian Ministry of health and population (No: 3-202027). The subjects were recruited from governmental hospitals and the participants and parents of young participants provided us with a written informed consent, as per the Helsinki Declaration on Human Experimentations guidelines.
Biochemical and anthropometric measurements
Anthropometric measurements such as body weight, waist circumference (WC), height, and hip circumference (HC) were measured by the regular ways [16], The body mass index (BMI) was calculated by dividing the weight in kilograms by height in meters squared (kg/m2), fasting serum triglycerides (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), very low-density lipoprotein cholesterol (VLDL-C), and glycelated hemoglobin (HbA1c) were measured enzymatically by using commercially available kits.
Genotyping assessment
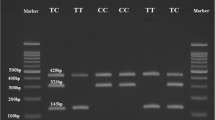
Blood samples were obtained from all the subjects in tubes with ethylenediaminetetraacetic acid anticoagulant. Total genomic DNA was extracted from the blood of all subjects using (PREP-RAPID, DNA, extraction kit, by (DNA-technology research and production ) company, Moscow, Russia) according to the manufacturer’s protocol, and to guarantee the quality of the extracted DNA concentrations were measured with Nano Drop (Nano Drop 2000/2000c, Thermo Scientific™). Analysis for the rs7574865 polymorphism was accomplished by using Taqman-probes-based real-time PCR reactions. This analysis was done in a 25-μL reaction with 20 ng of DNA, which was added to 12.5 μL TaqMan Universal Master Mix and 1.25 μL working stock. The samples were re-genotyped to endorse the results of the research. We used the Applied Biosystems Step One Real-time PCR - SDS version 2 software.
Statistical analysis
Genotypes were calculated by direct counting. We calculated Hardy–Weinberg equilibrium (HWE) by the chi-squared (χ2) test with expected frequencies derived from the actual frequencies in the SNP of patients and control. Anthropometric measurements and biochemical parameters between genotypes were compared using Student’s t test. The χ2 to compare between genotype frequencies for patients and control was done by SPSS software for windows (version 16.0; SPSS Inc, Chicago, IL). Moreover, the Hardy-Weinberg equilibrium status of patients and controls were measured by the goodness-of-fit chi-square test. Statistical significance was set at a probability (P value < 0.05).
Results
STAT4 genotype (rs7574865 G>T) frequencies were investigated in all 200 subjects (100 type 1 diabetes patients and 100 controls) as presented in (Table 1). It has been shown that the GT genotype has significantly higher frequency among T1D patients (75% GT and 2%TT, P < 0.05), than in control (30.0%) who mostly had wild-type GG genotype, Similarly, the minor allele (T) was most common among patients with percentage (41.3%) in comparison to control (15%) and (p = 0.01). Thus the outcomes suggest an association between the GT genotype and type 1 diabetes patients.
As demonstrated in Table 2, the demographics and clinical parameters of the study patients, subjects with the T allele, showed higher mean values for weight, Body mass index, waist circumference, hip circumference, glyclated hemoglobin, triglycerides and low-density lipids, and inferior value of high-density lipids and waist-hip circumference ratio. Furthermore, we assessed the correlation between the STAT4 rs7574865 variant and previously mentioned metabolic and anthropometric parameters, Table 3 showed that there was an association with HbA1C and TG with p value < 0.05.Álso the observed genotype frequencies of the SNP was statistically consistent with the expected distributions according to Hardy–Weinberg equilibrium.
Discussion
Type 1 diabetes incidence is due to multiple factors one of the most important factors is genetics, STAT proteins are cytosolic proteins that have a role in the intracellular signaling downstream of the type I and type II cytokine receptors [17,18,19], former studies and researches showed that type 1 diabetes incidence have been related to different genetic variants among different genes, such as a study on Tunisians showed that a variant of CREM gene, PTPN22, TCRβ, CD3z, ZAP70, and CTLA-4 gene variants correlation with T1D [20,21,22]. Moreover, it has been confirmed by earlier studies the correlation between the c.49A>G polymorphism of the CTLA4 gene and Egyptian, Lebanese, and Iraqi population and type 1 diabetes incidence [23,24,25]. Also, another prior study on the STAT4 gene showed confirmed the association of the STAT4 gene and various autoimmune diseases [26, 27] as the STAT4 gene codes a transcription factor that transfers signals induced by type 1 cytokines type1-IFN, IL-12, and IL-23 [28,29,30]. Moreover, it has a crucial role in the differentiation of T cells into the Th1. An earlier study on mice showed that diabetes type 1 was reduced by the interruption of the STAT4 signaling pathway [31]. And it has been proven earlier that some SNPs at STAT 4 gene contribute to the predisposition of diabetes type 1. This study support that the polymorphism rs7574865 has an association with the incidence of diabetes type 1 among Egyptians. rs7574865 variant, specifically, has been stated to be accompanied by various autoimmune diseases [15], and a preceding study confirmed that there is an association between rheumatoid arthritis and rs7574865 (T) allele among Spanish, Swedish, and Dutch patients and Egyptians [32, 33]. Also, it was proven its association with autoimmune thyroid diseases among the Chinese Han population [34]. Moreover, it was proven the susceptibility with AITD in the Asian population due to STAT4 rs7574865 polymorphism, but not in the African population [35]. Also, it is correlated with the susceptibility of systemic sclerosis [36], and colonic Crohn’s disease [37]. Furthermore, another study confirmed its association among Crete [38], and Mexican populations as well [39]. Also, there was a study that proved rs7574865 variant correlation with lupus among Asian population such as Japanese [40] and Chinese Northern Han population [41], and it was correlated as well with Egyptian population [42]. Most of the earlier studies on Asian and European populations confirmed that rs7574865 has a role in the incidence of diabetes type 1 [43, 44]. Also, a meta-analysis study performed on Caucasians and Asian subjects showed that it was associated with diabetes risk (P < 0.5) [45], and there was a study that confirmed the STAT4 was overexpressed from T1D Polish patients [46]. In contrast, a study among the Korean population, the Tunisian and Brazilian population showed no evidence of the association of the variant and T1D [20, 47, 48], while our study showed that the rs7574865 (T) allele is accompanied with increased risk for type 1 diabetes among Egyptians, same as another study on Greek patients [49] and Northeastern Chinese Han population [41, 44]. And we proved that the STAT4 (rs7574865) variant has a significant association and type 1 diabetes, and we found that the (GT) genotype is the most frequent among patients in comparison to the GG genotype as shown previously in (Table 1). We conducted this study because there were almost no studies among Africans and few among the Middle East area specifically the Arab region that shows the correlation between rs7574865 and type 1 diabetes.
Conclusion
This study showed that there was an association between rs7574865 polymorphism and f type 1 diabetes, these results could help to identify the mechanisms of type 1 diabetes well. And it’s highly recommended that more research should be done in depth to detect the genetics of T1D for treatment goals and providing novel ways for developing beneficial diagnostic and prognostic data for individuals. Moreover, more studies on other autoimmune disorders in Egyptian patients to determine if that variant is related to T1D or immune-related diseases.
Availability of data and materials
Not applicable
Abbreviations
- T1D:
-
Type 1 diabetes
- IL:
-
Interleukin
- STAT:
-
Signal transducer and activator of transcription
- PCR:
-
Polymerase chain reaction
- HbA1c:
-
Hemoglobin A1C
- LDL:
-
Low-density lipoprotein cholesterol
- HDL:
-
High-density lipoprotein cholesterol
- VLDL:
-
Very low-density lipoprotein cholesterol
- WC:
-
Waist circumference
- HC:
-
Hip circumference
- BMI:
-
Body mass index
- CTLA-4:
-
Cytotoxic T-lymphocyte-associated protein 4
- PTPN22:
-
Protein tyrosine phosphatase non-receptor type 22
- IFIH1:
-
Interferon induced with helicase C domain 1
- HLA:
-
Human leukocyte antigen
- TNF:
-
Tumor necrosis factor
- HWE:
-
Hardy–Weinberg equilibrium
References
Borchers AT, Uibo R, Gershwin ME (2010) The geoepidemiology of type 1 diabetes. Autoimmun Rev 9(5):A355–A365. https://doi.org/10.1016/j.autrev.2009.12.003
Federation ID (2017) IDF diabetes atlas 8th edition. International Diabetes Federation, pp 905–911
Harrington LE, Mangan PR, Weaver CT (2006) Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol 18(3):349–356. https://doi.org/10.1016/j.coi.2006.03.017
Donaghue KC, Chiarelli F, Trotta D, Allgrove J, Dahl-Jorgensen K (2009) Microvascular and macrovascular complications associated with diabetes in children and adolescents. Pediatr Diabetes 10:195–203. https://doi.org/10.1111/j.1399-5448.2009.00576.x
Goyal R, Jialal I (2020) Diabetes Mellitus Type 2. StatPearls. StatPearls Publishing, Treasure Island
Størling J, Pociot F (2017) Type 1 diabetes candidate genes linked to pancreatic islet cell inflammation and beta-cell apoptosis. Genes 8(2):72. https://doi.org/10.3390/genes8020072
Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, Babu SR, Eisenbarth GS (2006) Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci 103(38):14074–14079. https://doi.org/10.1073/pnas.0606349103
Braun L (2002) Race, ethnicity, and health: can genetics explain disparities? Perspect Biol Med 45(2):159–174. https://doi.org/10.1353/pbm.2002.0023
Lonard WJ, O'Shea JJ (1998) Jaks and STATs: biological implications. Annu Rev Immunol 16(1):293–322. https://doi.org/10.1146/annurev.immunol.16.1.293
Visconti R, Gadina M, Chiariello M, Chen EH, Stancato LF, Gutkind JS, O'Shea JJ (2000) Importance of the MKK6/p38 pathway for interleukin-12–induced STAT4 serine phosphorylation and transcriptional activity. Blood 96(5):1844–1852. https://doi.org/10.1182/blood.V96.5.1844
Decker T, Kovarik P (2000) Serine phosphorylation of STATs. Oncogene 19(21):2628–2637. https://doi.org/10.1038/sj.onc.1203481
Trembleau S, Penna G, Bosi E, Mortara A, Gately MK, Adorini L (1995) Interleukin 12 administration induces T helper type 1 cells and accelerates autoimmune diabetes in NOD mice. J Exp Med 181(2):817–821. https://doi.org/10.1084/jem.181.2.817
Rothe H, O'Hara RM, Martin S, Kolb H (1997) Suppression of cyclophosphamide induced diabetes development and pancreatic Th1 reactivity in NOD mice treated with the interleukin (IL)-12 antagonist IL-12 (p40) 2. Diabetologia 40(6):641–646. https://doi.org/10.1007/s001250050728
Rothe H, Burkart V, Faust A, Kolb H (1996) Interleukin-12 gene expression is associated with rapid development of diabetes mellitus in non-obese diabetic mice. Diabetologia 39(1):119–122. https://doi.org/10.1007/BF00400422
Martinez A, Varade J, Marquez A, Cenit MC, Espino L, Perdigones N et al (2008) Association of the STAT4 gene with increased susceptibility for some immune-mediated diseases. Arthritis Rheum 58(9):2598–2602. https://doi.org/10.1002/art.23792
Kroke A, Bergmann MM, Lotze G, Jeckel A, Klipstein-Grobusch K, Boeing H (1999) Measures of quality control in the German component of the EPIC study. Ann Nutr Metab 43(4):216–224. https://doi.org/10.1159/000012788
Schindler C, Levy DE, Decker T (2007) JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282(28):20059–20063. https://doi.org/10.1074/jbc.R700016200
Stark GR, Darnell JE Jr (2012) The JAK-STAT pathway at twenty. Immunity 36(4):503–514. https://doi.org/10.1016/j.immuni.2012.03.01
O’Shea JJ, Plenge R (2012) JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 36(4):542–550. https://doi.org/10.1016/j.immuni.2012.03.014
Zouidi F, Bouzid D, Fourati H, Fakhfakh R, Kammoun T, Hachicha M, Masmoudi H (2017) CREM variant rs17583959 conferred susceptibility to T1D risk in the Tunisian families. Immunol Lett 181:1–5. https://doi.org/10.1016/j.imlet.2016.11.007
Benmansour J, Stayoussef M, Al-Jenaidi FA, Rajab MH, Rayana CB, Said HB et al (2010) Association of single nucleotide polymorphisms in cytotoxic T-lymphocyte antigen 4 and susceptibility to autoimmune type 1 diabetes in Tunisians. Clin Vaccine Immunol 17(9):1473–1477. https://doi.org/10.1128/CVI.00099-10
Ferjeni Z, Bouzid D, Fourati H, Stayoussef M, Abida O, Kammoun T, Hachicha M, Penha-Gonçalves C, Masmoudi H (2015) Association of TCR/CD3, PTPN22, CD28 and ZAP70 gene polymorphisms with type 1 diabetes risk in Tunisian population: family based association study. Immunol Lett 163(1):1–7. https://doi.org/10.1016/j.imlet.2014.11.005
Mosaad YM, Auf FA, Metwally SS, Elsharkawy AA, El-Hawary AK, Hassan RH, Tawhid ZE, El-Chennawi FA (2012) HLA-DQB1* alleles and genetic susceptibility to type 1 diabetes mellitus. World J Diabetes 3(8):149–155. https://doi.org/10.4239/wjd.v3.i8.149
Wafai RJE, Chmaisse HN, Makki RF, Fakhoury H (2011) Association of HLA class II alleles and CTLA-4 polymorphism with type 1 diabetes. Saudi J Kidney Dis Transpl 22(2):273
Ahmed NS (2014) The codon 17 polymorphism of the CTLA4 gene in type 1 diabetes mellitus in the Baghdad population. J Med Genet Genom 6(1):1–5
Kobayashi S, Ikari K, Kaneko H, Kochi Y, Yamamoto K, Shimane K, Momohara S (2008) Association of STAT4 with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Japanese population. Arthritis Rheum 58(7):1940–1946. https://doi.org/10.1002/art.23494
Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, Criswell LA (2008) Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet 4(5):e1000084. https://doi.org/10.1371/journal.pgen.1000084
Korman BD, Kastner DL, Gregersen PK, Remmers EF (2008) STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep 8(5):398–403. https://doi.org/10.1007/s11882-008-0077-8
Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O’Malley JT, Kapur R, Levy DE, Kansas GS, Kaplan MH (2007) Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol 178(8):4901–4907. https://doi.org/10.4049/jimmunol.178.8.4901
Zervou MI, Goulielmos GN, Castro-Giner F, Tosca AD, Krueger-Krasagakis S (2009) STAT4 gene polymorphism is associated with psoriasis in the genetically homogeneous population of Crete, Greece. Hum Immunol 70(9):738–741. https://doi.org/10.1016/j.humimm.2009.05.008
Holz A, Bot A, Coon B, Wolfe T, Grusby MJ, von Herrath MG (1999) Disruption of the STAT4 signaling pathway protects from autoimmune diabetes while retaining antiviral immune competence. J Immunol 163(10):5374–5382
Orozco G, Alizadeh BZ, Delgado-Vega AM, González-Gay MÁ, Balsa A, Pascual-Salcedo D, Martín J (2008) Association of STAT4 with rheumatoid arthritis: a replication study in three European populations. Arthritis Rheum 58(7):1974–1980. https://doi.org/10.1002/art.23549
Settin A, Salama A, Elshazli R (2014) Signal transducer and activator of transcription 4 (STAT4) G>T gene polymorphism in Egyptian cases with rheumatoid arthritis. Hum Immunol 75(8):863–866. https://doi.org/10.1016/j.humimm.2014.06.013
Yan N, Meng S, Zhou J, Xu J, Muhali FS, Jiang W et al (2014) Association between STAT4 gene polymorphisms and autoimmune thyroid diseases in a Chinese population. Int J Mol Sci 15(7):12280–12293. https://doi.org/10.3390/ijms150712280
Gao X, Wang J, Yu Y (2019) The association between STAT4 rs7574865 polymorphism and the susceptibility of autoimmune thyroid disease: a meta-analysis. Front Genet 9:708. https://doi.org/10.3389/fgene.2018.00708
Dieudé P, Guedj M, Wipff J, Ruiz B, Hachulla E, Diot E, Allanore Y (2009) STAT4 is a genetic risk factor for systemic sclerosis having additive effects with IRF5 on disease susceptibility and related pulmonary fibrosis. Arthritis Rheum 60(8):2472–2479. https://doi.org/10.1002/art.24688
Glas J, Seiderer J, Nagy M, Fries C, Beigel F, Weidinger M, Brand S (2010) Evidence for STAT4 as a common autoimmune gene: rs7574865 is associated with colonic Crohn's disease and early disease onset. PLoS One 5(4):e10373. https://doi.org/10.1371/journal.pone.0010373
Zervou MI, Sidiropoulos P, Petraki E, Vazgiourakis V, Krasoudaki E, Raptopoulou A, Goulielmos GN (2008) Association of a TRAF1 and a STAT4 gene polymorphism with increased risk for rheumatoid arthritis in a genetically homogeneous population. Hum Immunol 69(9):567–571. https://doi.org/10.1016/j.humimm.2008.06.006
Ramírez OB, Rincón JM, Cobos RB, Ávila IA, Bello JR (2016) STAT4 confers risk for rheumatoid arthritis and systemic lupus erythematosus in Mexican patients. Immunol Lett 175:40–43. https://doi.org/10.1016/j.imlet.2016.05.003
Kiyohara C, Washio M, Horiuchi T, Tada Y, Asami T, Ide S, Takahashi H (2009) Cigarette smoking, STAT4 and TNFRSF1B polymorphisms, and systemic lupus erythematosus in a Japanese population. J Rheumatol 36(10):2195–2203. https://doi.org/10.3899/jrheum.090181
Li C, Zhao L, Wang W, Li H, Meng X, Chen S (2015) STAT4 rs7574865 polymorphism contributes to the risk of type 1 diabetes: a Meta analysis. Int J Clin Exp Med 8(2):2471–2475
El-Lebedy D, Raslan H, Ibrahim A, Ashmawy I, Abd El-Aziz S, Mohammed AM (2017) Association of STAT4 rs7574865 and PTPN22 rs2476601 polymorphisms with rheumatoid arthritis and non-systemically reacting antibodies in Egyptian patients. Clin Rheumatol 36(9):1981–1987. https://doi.org/10.1007/s10067-017-3632-7
Liang YL, Wu H, Shen X, Li PQ, Yang XQ, Liang L, Tian WH, Zhang LF, Xie XD (2012) Association of STAT4 rs7574865 polymorphism with autoimmune diseases: a meta-analysis. Mol Biol Rep 39(9):8873–8882. https://doi.org/10.1007/s11033-012-1754-1
Yuan H, Feng JB, Pan HF, Qiu LX, Li LH, Zhang N, Ye DQ (2010) A meta-analysis of the association of STAT4 polymorphism with systemic lupus erythematosus. Mod Rheumatol 20(3):257–262. https://doi.org/10.1007/s10165-010-0275-9
Yi J, Fang X, Wan Y, Wei J, Huang J (2015) STAT4 polymorphisms and diabetes risk: a meta-analysis with 18931 patients and 23833 controls. Int J Clin Exp Med 8(3):3566–3572
Fichna M, Żurawek M, Bogusz-Górna K, Małecki PP, Niechciał E, Sidoruk A, Furman K, Fichna P (2020) STAT4 sequence variant and elevated gene expression are associated with type 1 diabetes in Polish children. Cent Eur J Immunol 45(1):22–28. https://doi.org/10.5114/ceji.2019.92492
Park Y, Lee HS, Park Y, Min D, Yang S, Kim D, Cho B (2011) Evidence for the role of STAT4 as a general autoimmunity locus in the Korean population. Diabetes Metab Res Rev 27(8):867–871. https://doi.org/10.1002/dmrr.1263
de Azevêdo Silva, J., Tavares, N. A. C., Santos, M. M. S., Moura, R., Guimarães, R. L., Araújo, J., Crovella S & Brandão, L. A. C. (2015). Meta-analysis of STAT4 and IFIH1 polymorphisms in type 1 diabetes mellitus patients with autoimmune polyglandular syndrome type III.
Zervou MI, Mamoulakis D, Panierakis C, Boumpas DT, Goulielmos GN (2008) STAT4: a risk factor for type 1 diabetes? Hum Immunol 69(10):647–650. https://doi.org/10.1016/j.humimm.2008.07.004
Acknowledgements
Not applicable
Funding
This work did not receive funding from any organization or institution.
Author information
Authors and Affiliations
Contributions
NA has done the biochemical and anthropometric analysis. SS and SI have done the genotyping, DNA extraction, and the real-time PCR. ME has done the statistical analysis. MA, DS, and SS had a major role in the writing and revision of the manuscript and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This research was accepted by the Ethical Committee of the Egyptian Ministry of health and population (No: 3-202027). The participants and parents of young participants provided a written informed consent, as per the Helsinki Declaration on Human Experimentations guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdelmajed, S.S., El-Dessouky, M.A., SalahElDin, D.S. et al. Assessing the association of rs7574865 STAT4 gene variant and type 1 diabetes mellitus among Egyptian patients. J Genet Eng Biotechnol 19, 112 (2021). https://doi.org/10.1186/s43141-021-00214-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-021-00214-2