Abstract
Background
Over the past decade, various research studies have proved the interconnection between the inflammatory pathways and diabetes complication in clinical condition. The present study evaluated the anti-inflammatory and antioxidant activity. Further, the sample was tested for its pharmacokinetics properties and the best compounds were docked with the diabetic markers (DPP IV (PDB-ID: IJ2E) and SGLT2 (PDB-ID: 7VSI)).
Results
C.verticillata showed a good hydrogen peroxide (78.3 ± 0.34%, IC50 = 287.81 µg/ml) and superoxide scavenging activity (52.7 ± 1.26%, IC50 = 796.15 µg/ml). In addition, the sample was checked for its anti-inflammatory activity with protein denaturation (57.4 ± 0.19%, IC50 = 471.5 µg/ml) and proteinase inhibition assay (68.3 ± 0.48%, IC50 = 213.42 µg/ml). Further, the bioactive compounds detected from HPLC-ESI-MS/MS analyzed sample were checked for its drug likeliness by checking its ADME properties and toxicological parameters. It has been observed that except Loliolide, all the other compounds have followed the physicochemical parameters and proved to exhibit drug likeliness characteristics. The bioactive compounds that follow the Lipinski’s rule were taken further for in silico molecular docking analysis with the diabetic protein markers (DPP IV and SGLT2). Docking results revealed that Pyro pheophorbide a with DPP IV and Dihydromonacolin L acid with SGLT2 have recorded a maximum docking score of (− 9.4 kcal/mol) and (− 9.2 kcal/mol), respectively.
Conclusion
The observed results suggest that the identified and selected bioactive compounds from C.verticillata can be considered as a potential target molecule for the management of oxidative stress induced diabetic condition. Furthermore, the study also provides an insight on the effectiveness of the compounds on reducing the inflammation as well.
Similar content being viewed by others
Background
Inflammation is characterized as a host defense mechanism which involves the activation of immune system to remove the foreign antigen from the host system. The visual interpretations of inflammation include swelling, increased temperature at the site of injury, pain and redness [1]. It is achieved by the enhanced production of reactive oxygen species by the action of inflammatory cells [2], where it aids in the removal of invasive microorganisms. Nevertheless, prolonged production of ROS (reactive oxygen species) leads to oxidative stress and illness leading to chronic inflammation. Moreover, chronic inflammation leads to various kinds of illness like cancer, neurological diseases, diabetes, cardiovascular diseases, and so on.
According to [3], oxidative stress may be a major factor in the pathophysiology of diabetes-related macrovascular and microvascular complications which end up inflammation. The emergence of endothelial dysfunction is a warning sign of such damage. Numerous evidences are there to support the relationship between the immune and metabolic systems. It has been proved that the inflammatory pathways are the primary mediators of diabetes [3]. Research findings had already revealed the interconnection between diabetes and inflammation, where the mediators of inflammation like IL-6 and CRP (C-reactive protein and plasminogen activator inhibitor-1) have shown to be elevated in different diabetic conditions [4].
Increasing evidences propose that prolonged activation of pro-inflammatory pathways in the action of insulin might contribute to metabolic disorders like T2D (type 2 diabetes) [5]. Currently existing treatment strategies for type 2 diabetes mellitus, such as insulin delivery and oral anti-diabetic medications, are either ineffective or have negative side effects. Hence, it is critical to continue looking for an effective treatment that can assist people with type 2 diabetes and eventually cure them. Therefore, there is an increased demand in search for the new marine algae-based bioactive compounds with minimum or no side effects to the patients. Hence, the present study focuses on the search of novel bioactive compounds for the effective management of type 2 diabetes that has an effect on reducing oxidative stress induced inflammation. In addition, our work also focuses on the usage of computational tools to verify the ADMET (absorption, distribution, metabolism, excretion and toxicity) properties of the bioactive compounds as well as the in silico molecular docking analysis with the diabetic protein markers (DPP IV (Dipeptidyl peptidase) and SGLT2 (sodium-glucose cotransporter-2). This work will open the way for the use of marine bioactive compounds as an alternative in the curative treatment of diabetes.
Methods
The collection of four green marine algae (Chaetomorpha crassa, Caulerpa racemosa, Caulerpa verticillata and Caulerpa scalpelliformis) from Gulf of Mannar (9°28′N, 79°18′E) and extraction of the marine algal extract were followed as described by [6]. Briefly, the algal samples were collected and extracted for its phytochemicals and the antioxidant potential of the algal extracts was assessed based on three different antioxidant assay (ABTS, metal chelation and phosphomolybdenum assay). Moreover, the samples were also subjected to antidiabetic assays (α-amylase and α-glucosidase enzyme inhibition). Among the four algal extracts, C.verticillata showed a good antioxidant and anti-inflammatory activities. Hence, further analysis was carried out only with C.verticillata.
In vitro antioxidant assays
Hydrogen peroxide scavenging assay
The ability of the ethanolic extract of C.verticillata to scavenge hydrogen peroxide was measured by following the protocol using ascorbic acid as a standard [7]. To 1 ml of extract of varying concentration (100–800 µg/ml), 600 µl of 2 mM H2O2 prepared in phosphate buffer (pH 7.4) was added. The reaction mixture was completely mixed and incubated for 10 min at room temperature, and the absorbance was measured at 230 nm in UV–Vis spectrophotometer (1650 Shimadzu, Japan). The same protocol was followed for the standard ascorbic acid, the scavenging potential was calculated, and the percentage of inhibition was calculated using the following equation:
Abs T = Absorbance of test sample; Abs C = Absorbance of control.
Superoxide radial scavenging assay
The superoxide radical scavenging activity was measured based on the protocol followed by [8]. The superoxide radicals were generated by adding 500 µl of NBT (nitro blue tetrazolium) (0.3 mM), 500 µl of NADH (nicotinamide adenine dinucleotide) (0.936 mM), 500 µl of Tris HCL buffer (16 mM, pH 8.0) and 1 ml of algal extract with increasing concentration (100-800 µg/ml). The reaction was initiated by the addition of PMS (phenylmethanesulfonic acid) (0.12 mM), and the tubes were incubated at 25 °C for 5 min. The absorbance was measured at 560 nm. A similar treatment was given to the standard ascorbic acid, and the inhibition percentage was expressed using the following equation:
Abs T = Absorbance of test sample; Abs C = Absorbance of control.
In vitro anti-inflammatory assays
Protein denaturation assay
Denaturation of protein was assessed by the protocol followed by [9]. To 1 ml of 1% BSA (bovine serum albumin), 1 ml of marine algal extracts (100–800 µg/ml) and 1 ml of phosphate buffer saline (pH 6.4) were added. Once the reaction mixture was mixed, the tubes were incubated at 70 °C in water bath for 10–15 min. The tubes were allowed to cool, and the turbidity was measured in UV–Vis spectrophotometer at 660 nm. The protein denaturation inhibition percentage was calculated according to the formula as follows:
Abs T = Absorbance of test sample; Abs C = Absorbance of control.
Proteinase inhibitory assay
The proteinase inhibition was measured as described by [10]. To 1 ml of 20 mM Tris–Hcl buffer (pH 7.4), 0.06 mg of trypsin and 1 ml of algal extract (100–800 µg/ml) were added. The reaction mixture was vortexed completely and incubated for 30 min at 37 °C. Furthermore, 1 ml of 0.8% casein (w/v) was added to stop the reaction, and the tubes were centrifuged for 5 min at 2500 rpm. The absorbance of the supernatant was measured at 210 nm. The inhibition percentage was calculated by the following equation:
Abs T = Absorbance of test sample; Abs C = Absorbance of control.
HPLC–UV-ESI MS/MS analysis of C.verticillata
The HPLC-MS (high-pressure liquid chromatography–mass spectrometry) system consisted of a MS pump, autosampler, UV detector, and a triple-quadrupole, ESI QTOF high-resolution mass spectrometer (Bruker, USA) with Met Frag software for data acquisition and analysis. The sample separation was carried out using Agilent poroshell C18 reverse phase column (150 mm×4.6 mm), 2.7 µm particle size (part number: 683975-902), protected with a security guard cartridge (Zorbax eclipse, C8, 4.6×12.5 mm, 5µ). The sample was analyzed according to the standard protocol with some minor modifications [11]. Two different mobile phases were used, like Solvent A: water (with 0.1% formic acid), and Solvent B: acetonitrile (with 0.1% formic acid), with a column temperature of 25˚C and UV detector at 280nm. The flow rate was 300µl/min, and 10µl of sample was injected. The samples eluting out from UV detector were then directed to triple-quadrupole tandem mass spectrometer with an electrospray interface (ESI), operating in complete scan mode from m/z (50–2200). Mass spectra were acquired in positive modes with capillary voltage at 4500 V, end plate offset 500 V, nebulizer gas at 60 psi, dry gas 12.0L/min, and capillary dry temperature at 220 °C. The identification was achieved and processed using Met Frag workstation software.
ADME analysis and pharmacokinetics toxicity prediction analysis
The canonical structures of the four aforementioned compounds from HPLC-ESI-MS/MS analysis were retrieved from Pubchem database (www.pubchem.ncbi.nlm.nih.gov). The selected compounds were subjected to ADME analysis. For this, the compounds were analyzed for its physicochemical limitations, pharmacokinetics investigation and drug-likeness of the compounds with use of SWISSADME (http://www.swissadme.ch/index.php). Furthermore, the toxicity prediction for the oral consumption was also calculated using Protox ii (tox.charite.de). This toxicity prediction helps in evaluating the carcinogenicity, hepatotoxicity, immunotoxicity and also the toxicological pathways [12].
In silico molecular docking analysis
Preparation of ligands and protein molecule
The crystal structures of the selected bioactive compounds (ligands) of C.verticillata were downloaded from Pubchem database. The 3D SDF structure of DPP IV (PDB-ID: IJ2E) and SGTL2 (PDB-ID: 7VSI) protein was retrieved from the protein data bank (www.rcsb.org). The protein was carefully validated using Ramachandran plot before the docking process using MolProbity server [13]. Further, the protein molecules were prepared by removing the water molecules, adding hydrogen atoms, removing the heteroatoms and bounded other polypeptide inhibitors. The modified protein molecule was used for the docking study [12].
In the current study, PyRx 0.8 version (https://pyrx.sourceforge.io/) software was used for docking purpose, which included Open babel, Auto and AutoDock Vina. Furthermore, the protein and the ligand molecules were transformed to pdbqt format using Autodock tools. Open babel was used to minimize the energy, and blind docking was performed. For a promising protein–ligand binding, the active binding regions were recognized using Biovia discovery studio tool. Analysis of the docked proteins is performed once docking is complete and visualized by BioVia, Discovery studio, 2020. Further, the inhibition constant was calculated to find out the binding efficacy of the ligand and protein molecule using the following equation [14]
where R = 1.985 × 10–3 kcal/mol (universal gas constant; T = 298.15 K (temperature);
ΔG = Binding energy.
Statistical analysis
All the tests were carried out in triplicate, and the data were represented as mean ± SE. The experimental results were statistically analyzed using GraphPad prism 9 and Microsoft Excel 2010.
Results
The ability of the ethanolic extract of C.verticillata has shown that the extract has a strong ability to scavenge hydrogen peroxide radicals at a dose-dependent manner. At a highest concentration of 800 µg/ml, C.verticillata extract has shown a maximum scavenging activity of 78.3 ± 0.34% with an IC50 of 287.8 µg/ml compared to the standard ascorbic acid (71.8 ± 1.15% at 200 µg/ml) (Table 1). The superoxide radical scavenging activity was determined using PMS-NADH method. The reduction in absorbance at 560 nm showed that the algal extract has the ability to quench the superoxide radicals, where the ethanolic extract of C.verticillata at a maximum concentration of 800 µg/ml has observed to show 52.7 ± 1.26% (IC50 = 796.15 µg/ml) when compared with the standard ascorbic acid (69.7 ± 1.02%, IC50 = 115.5 µg/ml) at 200 µg/ml.
Protein denaturation assay was carried out to measure the efficiency of the algal extracts to inhibit the denaturation of the proteins. C.verticillata has recorded a maximum inhibition of 57.4 ± 0.19% (IC50 = 471.5 µg/ml) with respect to the standard drug diclofenac of 96.4 ± 0.25% (IC50 of 9.4 µg/ml). In Table 2, the effect of C.verticillata extract on anti-proteinase activity is presented. The extract has recorded a good level of proteinase inhibition of about 68.3 ± 0.48% (IC50 = 213.42 µg/ml) with a significance of p < 0.05. The results have shown a good anti-proteinase activity in comparison to the standard aspirin (94.4 ± 0.22%, IC50 = 19.7 µg/ml). The results clearly indicate that the extract has the potential to bind with the cell surface by stabilizing the membrane with minor alterations on the charges of the cells.
C.verticillata was subjected to HPLC-ESI-MS/MS analysis for the identification of the bioactive compounds. It was identified as 2-Palmitoylglycerol, Pyro pheophorbide a, Dihydromonacolin and Loliolide are reported for its antioxidant and antidiabetic activities. The fragmentation of mass spectrum was carefully analyzed for its m/z values (Fig. 1). The chemical structure of the ESI-MS identified compounds is listed in Table 3. The compound peaked at 33.8 min retention time (Rt) was identified as Loliolide (C11H16O3) with a molecular ion peak at m/z 197.11 [M + H] +. Further, the compound at Rt = 56.6 min was identified as Dihydromonacolin L acid (C19H32O4) consisting of molecular ion peak at m/z 325.23 [M + H] +. The peaks at 59.0 min and 78.6 min were identified as 2-Palmitoylglycerol (C19H38O4) and Pyro pheophorbide a (C33H34N4O3) containing a molecular ion peak at m/z 331.28 [M + H] + and m/z 535.26 [M + H] +, respectively. Additionally, the fragmentation pattern of mass spectra was already reported in our previous published data [15]. The bioactive compounds after HPLC-ESI-MS/MS analysis were checked for its physicochemical properties like lipophilicity, solubility, bioavailability, number of hydrogen atoms, TPSA, BBB, and Lipinski rule; rotatable bonds were evaluated carefully using SWISSADME software tool. Firstly, Fig. 1 represents the bioavailability radar charts, which gives a quick glimpse of six important physicochemical properties like lipophilicity, size, polarity, flexibility and saturation. The compounds which fall under the pink area in radar charts refer that the compound comes under the property of drug likeliness (Fig. 2).
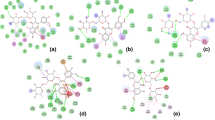
All the leading ligand molecules (2-Palmitoylglycerol, Loliolide, Pyro pheophorbide a and Dihydromonacolin L acid) were observed to follow maximum drug-likeness limitations. Moreover, all the compounds were observed to possess a good gastrointestinal absorption and solubility. Further, the compounds were subjected to toxicity prediction, where LD50 plays an important role in determining their level of toxicity, where it includes immunotoxicity, carcinogenicity, TPSA value and octanol/water partition (LogP) value (Table 4). All the four compounds were thoroughly evaluated for its pharmacochemical and physicochemical properties for in silico molecular docking analysis. The Ramachandran plot analysis of the protein (DPPIV) clearly revealed that most of the residues lie within the favored regions that indicate the protein quality (Fig. 3). Except Loliolide, docking was performed for all the other three compounds with the diabetic protein markers like DPP IV (PDB: IJ2E) and SGLT2 (PDB: 7VSI). Docking scores were assessed between the ligands and the proteins. The results revealed that Pyro pheophorbide a has a strong binding affinity with DPP IV (− 9.4 kcal/mol), followed by Dihydromonacolin L acid (− 7.7 kcal/mol) and 2-Palmitoylglycerol (− 5.3 kcal/mol) (Fig. 4a–f).
Discussion
The physiological and biological condition of the marine seaweeds that persist in varying conditions like temperature, pH, salinity and various other factors leads to the production of various phytoconstituents and potential bioactive compounds as a measure of defense mechanism. These seaweeds contain a variety if bioactive compounds that has the potential to quench the free radicals. It is well known that hydrogen peroxide is present everywhere in the environment at a low concentration. Earlier studies have reported that it is a weak oxidizing agent with the capability of crossing the cell membranes quickly and acts in response to Cu2+ and Fe2+ ions leading to the formation hydroxy radicals [16]. These hydroxyl radicals have the potential to initiate lipid peroxidation that leads to the damage of DNA. Hence, from the results it is very clear that the algal extract has the capability of scavenging hydrogen peroxide where the occurrence of potential phytoconstituents would have involved in electron donation to neutralize the radicals [17].
It is to be noted that enzymatic antioxidants like superoxide dismutase and catalase are involved in the decomposition of hydrogen peroxide to oxygen and water. Hence, changes in the enzymatic antioxidants due to hyperglycemic condition could lead to inactivation of superoxide dismutase where its glycosylation leads to decrease in activity [18]. Therefore, diabetes-induced oxidative stress increases the concentration of pro-inflammatory cytokines that results in the inflammation [19]. Many scientific reports had already proved that increased production of TNF-α has a strong link with obesity induced insulin resistance that leads to type 2 diabetes [20]. The present findings indicate that the extract has the potential to inhibit the denaturation of the protein by stabilizing the membrane of the protein. Various factors like heat, hypotonic environment, etc., can lead to membrane damage of the protein molecule that further results in the leakage of the inner lysosomal contents. The extracellular release of the membrane further leads to inflammation in the tissue. Hence, it is proved that the anti-inflammatory activity of C.verticillata extract provides an evidence for the stabilization of the membrane by inhibiting the discharge of lysosomal constituents [21].
Previous reports have evidenced that prolonged discharge of inflammatory intermediates would upregulate the expression of genes involved in acute and chronic inflammatory diseases by inactivating the anti-proteinases. Earlier reports of [22] have stated that uninhibited levels of proteolysis by the proteinase enzyme can result in various patho-physiological symptoms. The equilibrium of anti-proteinases and proteinases is to be maintained and regulated. Under stress conditions, the ROS will inactivate the anti-proteinases by which the balance gets disturbed resulting in uncontrolled damage of proteins causing increased tissue injury. Therefore, a natural remedy is required to have both anti-inflammatory and antioxidant property that can lessen the ROS-induced stress and regulate the immune function. Hence, C.verticillata could be the right choice to prevent the rupture of the HRBC membrane, by which the erythrocyte membrane gets stabilized. By stabilizing the lysosomal membrane, the inflammatory process gets regulated and prevents the release of inflammatory mediators. Our current results of C.verticillata have provided the evidence for the aforementioned activities.
Currently, many effective medications made from natural sources are discovered utilizing computer-aided drug design approaches because of the ongoing advancements in computer science. Nowadays, computer-aided drug design is employed to anticipate ADMET features of bioactive compounds, resulting in early stage drug development [23]. The motivation for this in silico techniques is due to the reduced cost and time factor required when compared to traditional ADMET profiling. With respect to drug development, it is important to consider the concept of inhibition constant (Ki) that helps to prioritize the bioactive compounds as efficient inhibitors for therapeutic purpose. The inhibition constant of the compounds is listed in Table 5 which is expected to lie between the ranges of 0.1–1.0 µM [13]. The molecular docking results clearly implies that there is a strong relationship with respect to the docking score (ΔG) and the inhibition constant (Ki). The inhibition constant was calculated according to the formula as mentioned in the methodology section (Eq. (1). The results revealed that the compounds that showed maximum binding energy (ΔG) (DPP IV: Pyro pheophorbide a (− 9.4 kcal/mol) possess a lower inhibition constant (Ki = 1.31 µM). From this, it is evident that higher binding energy and lower inhibition constant indicate a stronger and a stable ligand–protein interaction [24] since it provides a deeper insight about the compound to regulate the particular biological pathways. Also, these bioactive compounds were subjected to SWISS ADME software to evaluate the drug likeliness and the physicochemical properties of the compounds, toxicity prediction and in silico molecular docking. These preclinical compounds have the ability to get absorbed to the surface of the receptor that is again circulated into the target site. Further, these compounds will be metabolized by the liver and excreted out of the system to avoid toxicity [25]. In the present study, almost all the bioactive compounds have followed the pharmacochemical and physicochemical parameters (molecular weight: less than 500D), Log S which refers the solubility. The result showed that all the compounds except Loliolide were inactive for immunotoxicity and carcinogenicity, whereas Loliolide showed positive to carcinogenicity. Additionally, the LD50 value of Loliolide was 34 mg/kg; hence, it comes under class 2 of toxicity, which is fatal if swallowed.
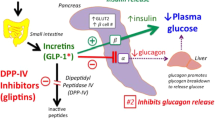
Considering the Ramachandran plot analysis, the widespread of ψ (psi) and φ (phi) angles clearly specifies the flexibility and potential ability of the protein to undergo conformational changes on binding of the ligand molecule [14]. These observations helped in accounting for the protein dynamics in molecular docking studies (Fig. 3). Except Loliolide, docking was performed for all the other three compounds with the diabetic protein markers like DPP IV (PDB: IJ2E) and SGLT2 (PDB: 7VSI). Docking scores were assessed between the ligands and the proteins. The results revealed that Pyro pheophorbide a has a strong binding affinity with DPP IV (− 9.4 kcal/mol), followed by Dihydromonacolin L acid (− 7.7 kcal/mol) and 2-Palmitoylglycerol (− 5.3 kcal/mol). Earlier reports suggest that DPP IV was considered as a target to treat type 2 diabetes by inactivating glucagon inhibitory peptide and GLP-1 (glucagon-like peptide) [26]. Additionally, DPP IV inhibition was suggested to show a beneficial effect on postprandial glucose levels [27]. Moreover, it is proved that Pyro pheophorbide has the ability to inhibit alpha amylase and lipase by which it decreases the absorption of triglycerides present in the intestine that prevents from hyperlipidemia and obesity [28]. It has been evidenced that administration of Pyro pheophorbide a has significantly reduced the blood glucose level in normal and diabetic mice by inhibiting alpha amylase and alpha glucosidase enzymes [29]. Similarly, 2-Palmitoylglycerol has the ability to activate GPR119 which is predominantly expressed in pancreatic-β cells, where its activation leads to induce the secretion of insulin and GLP-1 [30].
Since studies had shown that the incretin hormones, glucose-dependent insulin tropic polypeptide (GIP) and glucagon-like peptide-1 are important regulators of postprandial insulin production; inhibition of DPP4 might be a great choice of interest to treat type 2 diabetes. It is also proved that 2-Palmitoylglycerol has the potential of reducing the inflammation and additionally minimizes the level of fatty acid oxidation [31]. Hence, strong interaction of the ligands (2-Palmitoylglycerol, Pyro pheophorbide a and Dihydromonacolin L acid) with the active site of DPP IV protein proves that these compounds could be a great choice for the management of type 2 diabetes (Fig. 2a, c and e). SGTL2 (sodium–glucose co-transporters) plays a very important role in the reabsorption of glucose in the kidney. Inhibition of SGTL2 increases the excretion of glucose in the urine by decreasing the glucose level in the plasma [32]. Existing literature suggests that the primary method by which SGLT2 reduces inflammation and oxidative stress was its capacity to minimize the negative impact of an overabundance of nutrients on cells and tissues, especially on the adipose tissues. When adipose tissue is exposed to an overabundance of nutrients, it produces more pro-inflammatory hormones and cytokines) TNF, IL-1, IL-6 and leptins) [33].
In our present study, Dihydromonacolin L acid was shown to dock a maximum score of -9.2 kcal/mol with SGTL2 followed by Pyro pheophorbide a (− 8.7 kcal/mol) and 2-Palmitoylglycerol (− 6.6 kcal/mol), which directly implies that the compound has the ability to reduce the glucose level and lessen the effects of inflammation induced oxidative stress as well (Fig. 2b, d and f). Previously, it was reported that the Dihydromonacolin L acid was proved to be a potential inhibitor in the biosynthesis pathway of cholesterol [34]. Hence, it protects from LDL oxidation by inhibiting HMG-CoA reductase [35]. Pheophorbide is a by-product of chlorophyll which is known for its anti-inflammatory and anticancer property, where it inhibits the high concentration of nitric oxide (NO) to suppress the induction of iNOS (nitric oxide synthase) [36]. Hence, these bioactive compounds were expected to be a possible choice for the treatment and management of type 2 diabetes.
Conclusion
The ultimate aim of the drug discovery research is to find effective medications through clinical and preclinical studies. In this context, the integration of docking and ADMET studies has provided a deeper insight into the understanding about the ligand–protein interaction and pharmacokinetics of the tested compounds. The results of the present study conclude that C.verticillata has the potential to inhibit the upregulation of blood glucose level. Moreover, the extract has the ability to reduce oxidative stress induced inflammation as well. The bioactive compound extracted (2-Palmitoylglycerol, Dihydromonacolin L acid, Loliolide and Pyro pheophorbide a) from C.verticillata could be considered as one of the promising agents for treating T2D. Even though a number of bioassay experiments were performed for compound's to evaluate its ADMET profile, they are quite time-consuming and costly. This emphasizes on the importance of in silico predictions for its ADMET properties. Ideally, ADME, toxicity prediction and in silico analysis have revealed that the aforementioned bioactive compounds have a greater affinity to DPP4 and SGTL2. The outcome of the analysis suggests that the docked bioactive compounds were found to be non-hazardous and the computed drug analysis needs further validation to find its effectiveness in treating oxidative stress and inflammation in diabetic condition. To ensure the inhibitors of DPP4 and SGTL2 from marine algal source, the analyses necessitate further validation through in vivo experiments. We believe that expanding this research by integrating proper in vivo experiments along with ADMET and in silico analysis gave a deeper insight about the bioactive compounds from marine seaweeds as a potential therapeutic candidate in the treatment of oxidative stress induced diabetes. Hence, this holistic approach will eventually pave the way for the advancement in drug discovery to produce more safer and effective therapeutic agents.
Availability of data and materials
Research data are available upon request. Data can be obtained from the corresponding author via email.
Abbreviations
- DPP IV:
-
Dipeptidyl peptidase 4
- SGTL2:
-
Sodium-glucose cotransporter-2
- ROS:
-
Reactive oxygen species
- CRP:
-
C- reactive protein
- IL-6:
-
Interleukin 6
- T2D:
-
Type 2 diabetes
- ADMET:
-
Absorption, distribution, metabolism, excretion and toxicity
- ABTS:
-
2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid
- NBT:
-
Nitro blue tetrazolium
- NADH:
-
Nicotinamide adenine dinucleotide
- PMS:
-
Phenylmethanesulfonic acid
- BSA:
-
Bovine serum albumin
- HPLC-MS:
-
High-pressure liquid chromatography–mass spectrometry
- ESI-MS:
-
Electrons spray ionization–mass spectrometry
- BBB:
-
Blood–brain barrier
- TPSA:
-
Topological polar surface area
- R t :
-
Retention time
References
Marques-Vidal P, Schmid R, Bochud M (2012) Adipocytokines, hepatic and inflammatory biomarkers and incidence of type 2 diabetes. The CoLaus study. PLoS One 7:e51768
Anosike CA, Obidoa O, Ezeanyika LU (2012) The anti—Inflammatory activity of garden egg (Solanum aethiopicum) on egg albumin—Induced oedema and granuloma tissue formation in rats. Asian Pac J Trop Med 5:62–66
Oso BJ, Adeoye AO, Olaoye IF (2020) Pharmacoinformatics and hypothetical studies on allicin, curcumin, and gingerol as potential candidates against COVID-19-associated proteases. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1813630
Barriuso J, Nguyen DT, Li JW, Roberts JN, MacNevin G, Chaytor JL, Marcus SL, Vederas JC, Ro DK (2011) Double oxidation of the cyclic nonaketide Dihydromonacolin L to monacolin J by a single cytochrome P450 monooxygenase, LovA. J Am Chem Soc 133(21):8078–8081
Ben-Shabat S et al (1998) An entourage effect: inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur J Pharmacol 353(1):23–31
Bhatti MZ, Ali A, Ahmad A et al (2015) Antioxidant and phytochemical analysis of Ranunculus arvensis L. extracts. BMC Res Notes 8:279
Chu ZL, Carroll C, Chen R, Alfonso J, Gutierrez V, He H, Lucman A, Xing C, Sebring K, Zhou J (2010) N-Oleoyldopamine enhances glucose homeostasis through the activation of GPR119. Mol Endocrinol 24:161–170
Sakat S, Juvekar AR, Gambhire MN (2010) In vitro antioxidant and anti-inflammatory activity of methanol extract of Oxalis corticata Linn. Int J Pharm Pharm Sci 2:146–155
Duncan BB, Schmidt MI, Pankow JS (2003) Low-grade systemic inflammation and the development of type 2 diabetes: the atherosclerosis risk in communities study. Diabetes 52:1799–1805
Santer R (2003) Molecular analysis of the SGLT2 gene in patients with renal glucosuria. J Am Soc Nephrol 14:2873–2882
Endo A, Hasumi K, Nakamura T, Kunishima M, Masuda M (1985) Dihydromonacolin-L and monacolin-X, new metabolites those inhibit cholesterol-biosynthesis. J Antibiot 38:321–327
Ghosh AK, Gemma S (2015) Structure-based design of drugs and other bioactive molecules. John Wiley & Sons, Hoboken, pp 397–409
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21
Falade VA, Adelusi TI, Adedotun IO (2021) In silico investigation of saponins and tannins as potential inhibitors of SARS-CoV-2 main protease (Mpro). In Silico Pharmacol 9:9
Maheswari A, Salamun DE (2022) In vitro correlation studies of antidiabetic, antioxidant activity and HPLC-ESI-MS/MS analysis of marine seaweeds from Gulf of Mannar. Reg Stud Mar Sci 56:102682
Giugliano D, Ceriello A, Paolisso G (1996) Oxidative stress and diabetic vascular complications. Diabetes Care 19:257–267
Gunathilake KDPP, Ranaweera KKDS, Rupasinghe HPV (2018) Influence of boiling, steaming and frying of selected leafy vegetables on the in vitro anti-inflammation associated biological activities. Plants 7:22
Hurley JVJV (1972) Acute inflammation. Churchill Livingstone, Edinburgh
Islam MN, Ishita IJ, Jin SE, Choi RJ, Lee CM, Kim YS, Jung HA, Choi JS (2013) Anti-inflammatory activity of edible brown alga Saccharina japonica and its constituents pheophorbide a and pheophytin a in LPS-stimulated RAW 264.7 macrophage cells. Food Chem Toxicol 55:541–548
Istvan ES, Deisenhofer J (2001) Structural mechanism of statin inhibition of HMG CoA reductase. Science 29:1160–1164
Robak J, Gryglewski RJ (1988) Flavonoids are scavengers of superoxide anions. Biochem Pharmacol 37:837–841
Miller MJ, Sadowska-Krowicka H, Chotinaruemol S, Kakkis JL, Clark DA (1993) Amelioration of chronic ileitis by nitric oxide synthase inhibition. J Pharmacol Exp Ther 264(1):11–16
Kim MJ, Kim HJ, Han JS (2018) Pheophorbide A from Gelidium amansii improves postprandial hyperglycemia in diabetic mice through α-glucosidase inhibition. Phytother Res. https://doi.org/10.1002/ptr.6260
Abdul-Hammed M, Adedotun IO, Falade VA et al (2021) Target-based drug discovery, ADMET profiling and bioactivity studies of antibiotics as potential inhibitors of SARS-CoV-2 main protease (Mpro). VirusDis 32:642–656
Adelusi TI, Oyedele AQ, Boyenle ID, Ogunlana AT, Adeyemi RO, Ukachi CD, Idris MO, Olaoba OT, Adedotun IO, Kolawole OE, Xiaoxing Y, Abdul-Hammed M (2022) Molecular modeling in drug discovery. Inform Med Unlocked 29:100880
Packer M (2020) Mitigation of the adverse consequences of nutrient excess on the kidney: a unified hypothesis to explain the reno protective effects of sodium-glucose cotransporter 2 inhibitors. Am J Nephrol 51:289–293
Perseghin G, Petersen K, Shulman GI (2003) Cellular mechanism of insulin resistance: potential links with inflammation. Int J Obes Relat Metab Disord 27(Suppl. 3):S6–S11
Pieper GM (1998) Review of alterations in endothelial nitric oxide production in diabetes: protective role of arginine on endothelial dysfunction. Hypertension 31:1047–1060
Ruch RJ, Cheng SJ, Klaunig JE (1989) Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 10:1003–1008
Drucker DJ, Nauck MA (2006) The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 368:1696–1705
Verspohl EJ (2009) Novel therapeutics for type 2 diabetes: incretin hormone mimetics (glucagon-like peptide-1 receptor agonists) and dipeptidyl peptidase-4 inhibitors. Pharmacol Ther 124:113–138
Seven A, Guzel S, Seymen O, Civelek S, Bolayirli M, Uncu M, Burcak G (2004) Effects of vitamin E supplementation on oxidative stress in streptozotocin induced diabetic rats: investigation of liver and plasma. Yonsei Med J 45:703–710
Chatterjee S (2016) Oxidative stress, inflammation, and disease. In: Oxidative stress and biomaterials, pp. 35–58.
Shoelson SE, Lee J, Goldfine AB (2006) Inflammation and insulin resistance. J Clin Invest 116:1793–1801
Stockley RA (2000) Alpha-1-antitrypsin deficiency: what next? Thorax 55:614–618
Sugiyama H (2007) Oligomeric procyanidins in apple polyphenol are main active components for inhibition of pancreatic lipase and tri-glyceride absorption. J Agric Food Chem 55:5906–5906
Acknowledgements
The authors are thankful to the management of Jain (Deemed-to-be University) for providing required facilities for carrying out the research work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
SDE contributed to conceptualization, investigation and formal analysis; MA contributed to conceptualization, methodology, and original draft writing—review and editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
A., M., DE, S. Integrating in silico molecular docking, ADMET analysis of C.verticillata with diabetic markers and in vitro anti-inflammatory activity. Futur J Pharm Sci 10, 3 (2024). https://doi.org/10.1186/s43094-023-00576-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43094-023-00576-z