Abstract
Background
Surgical site infection (SSI) is the most typical problem for patients who undergo operative procedures. It remains a typical and widespread problem causing morbidity and mortality, partly related to a rise in infections due to antimicrobial-resistant bacterial pathogens. The study was purposed to evaluate the bacterial isolates and their drug susceptibility patterns in patients with postoperative surgical site infection.
Results
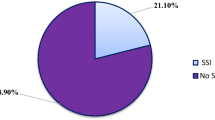
The rate of postoperative surgical site infections was studied at Asella Referral and Teaching Hospital and found to be 23.3%. One hundred fifty specimens of pus and surface swabs were collected from the surgical site-infected patients over the period of March 2016 to May 2017, and from that, a total of 147 bacterial pathogens were recovered. The predominant organisms associated with postoperative surgical site infections were Klebsiella species 38 (26%, n=147), Escherichia coli 31 (21%, n=147), Staphylococcus aureus 25 (17%, n=147), and Pseudomonas aeruginosa 18 (12%, n=147).
Conclusion
Higher number of bacterial isolates were recovered. The predominant isolates were Klebsiella species, Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. The majority of gram-negative bacterial species which were isolated were found to be resistant to the commonly prescribed antimicrobial agents in the study setting. Thus, to achieve effective therapy for wound infections and to reduce/stop the appearance of multidrug-resistant (MDR) pathogens, continuous monitoring is essential with the fair use of antimicrobial agents.
Similar content being viewed by others
Background
The hospital setting is a possible source of microbial infections as it lodges a large number of susceptible individuals and patients with various pathogenic microorganisms. In the hospital setting, the augmented prevalence of pathogenic microorganisms is correlated with a concurrent increase in different forms of nosocomial infections [1].
Surgical site infection (SSI) is characterized as an infection at the surgical site within 30 days after the surgery (or within 1 year if the implant is left in place), affecting either the incision or the deep tissue. These infections can involve apparent or deep tissue infections, organ or cavity infections [2]. Surgical site infections are the severe issues in the hospital population problems for patients undergoing surgeries, with the third most commonly identified hospital-acquired infection [2]. Intensified illness, extended hospitalization, death, and substantial financial burden for medical care are associated with postoperative surgical site infections [3, 4].
SSI prevention strategies have been highly developed, including enhanced ventilation of the operating theater, sterilization techniques, medical and surgical procedures, and the availability of prophylactic antibiotics. However, due to the proliferation of resistant pathogens, these SSIs are significant sources of morbidity and mortality [1]. The improper use of surgical prophylactic antibiotics leads partially to SSIs [3].
Antimicrobial resistance is a complex factor in controlling infections. It can increase complications and costs associated with procedures and treatment. An infected site complicates the postoperative course and results in a prolonged hospital stay and delayed recovery [5]. Advances in the treatment of diseases have contributed to a considerable rise in various surgical procedures. In response to these advancements, the severity of SSIs, antimicrobial prophylaxis, and the proliferation of antibiotic-resistant pathogens are expected to increase [5].
Certain life-saving medical procedures, like appendectomies and cesarean operations, are related to a high chance of infections and even mortality in underdeveloped countries, especially those with limited medical resources. Consequently, infections associated with the postoperative procedure usually increase the patient’s hospital stay by an average of 7 to 10 days, and in some instances, it may cause the death of the patient [6, 7].
For a long time, postoperative infections associated with surgery have been an established crisis. Researchers identified the various types of pathogens from the surgical wound in patients related to Ethiopia. These pathogens are Staphylococcus aureus and certain species associated with Klebsiella, Enterobacter, Proteus, Pseudomonas, Streptococcus, and coagulase-negative Staphylococcus (CoNS) [8, 9]. The rate of hospital-acquired infection is distinctly elevated in many developing countries like Ethiopia, particularly for mostly preventable infections (e.g., those following surgical operations). For example, in Mekelle Hospital, Ethiopia, the incidence of postoperative surgical site infection was 44.1% of the patients with hospital-acquired infection [10].
Due to antimicrobial-resistant infections and susceptibility trends to frequently prescribed medication used for management, minimal evidence is available about the severity of SSIs in Ethiopia. This disparity makes it more difficult for physicians to choose empirical therapy. So, in light of the above facts, the study was aimed to isolate, cultivate, and identify the bacteria from the surgical sites and to assess their susceptibility pattern for the commonly prescribed antibiotics in the study setting.
Methods
A prospective study was planned and conducted in a 321 bedded hospital, a public sector referral and teaching hospital in Ethiopia. The target departments for the study were general surgical and orthopaedic units. All the 643 individuals who underwent elective surgical interventions between March 2016 and May 2017 were assessed for surgical site infections, and the patients who were suspected of postoperative wound infections were enrolled in the study. The elective surgical procedures/interventions included laparotomy, appendectomy, prostatectomy, thyroidectomy, debridement, and incisions. The commonly performed surgeries under emergency conditions were exploratory laparotomy and appendectomy.
On the first day of patients’ admission to the hospital, the relevant information from the patient was collected by the surgeon. Besides, the surgical site was evaluated by a surgeon (who himself is one of the other operating surgeons) daily during the tenure of the hospital stay, two times a week up to 14 days postoperatively. However, for the observation of other complications, the surveillance time was extended up to 30 days after hospital discharge.
Two experienced clinical pharmacists used pretested structured questionnaires to obtain information from the patient’s case files. The information included were demographics (such as age, sex), existing chronic conditions (such as heart disease, metabolic syndrome), and previous medical condition (past diagnosis and cure like cataract surgery, dental restorations, and burn excision). Also, information like current medication, alcoholism, duration of preoperative stay in the hospital, duration of surgery, and, if given, antimicrobial prophylaxis and drain used were gathered using pretested structured questionnaires [11]. In the case of a suspected patient of wound infection, the wound site samples were collected as per the centers’ guidelines for disease control and prevention (CDC) [12]. The samples from the wound secretions were collected aseptically by the treating surgeons with the help of sterile cotton swabs without contaminating them with skin floras. All samples were received before cleaning the wound with antiseptic as well as before starting the antibiotic therapy for healing the wound.
Collected swab specimens were sent to the Asella Referral and Teaching Hospital’s microbiology laboratory for culture and sensitivity testing to identify, isolate, and understand the isolates’ susceptibility pattern against various antibiotics. Standard microbiological procedures like Gram staining, biochemical evaluation, and morphological examinations were applied for the bacterial isolation and identification and drug susceptibility evaluation. According to the standard guidelines, classical biochemical tests were performed for the species identification of the isolates from pure colonies [13]. The antibiotics (drugs) tested for susceptibility for both gram-negative and gram-positive bacteria were ampicillin (10μg), ciprofloxacin (5μg), gentamicin (10μg), chloramphenicol (30μg), doxycycline (30μg), ceftazidime, and ceftriaxone (30μg). Methicillin (5μg) and vancomycin (30μg) were used for only gram-positive bacterial isolates. Pre-analytical, analytical, and post-analytical stages of quality assurance incorporated in standard operating procedures (SOPs) of the Asella Referral and Teaching Hospital’s microbiology laboratory were strictly followed.
Ethical consideration
The Institutional Ethical Review Committee approved the study (Approval Reference Number: A/CHS/RC005/15/16). Written informed consent from each study participant was obtained. All the data collected from participants in the study was kept confidential. The study participant’s laboratory outcome was reported to the treating physicians for adequate care.
Statistical analysis
All data were evaluated statistically (Graph Pad Prism version 5.01), and mean ± SD was calculated for age. The comparison between the two groups was made by applying a chi-squared test. Significant differences were observed for the two groups based on age, sex, previous antibiotic exposure within 1 month, received antibiotic prophylaxis, and type of antibiotics prescribed for prophylaxis. All comparisons were made at P ≤ 0.05.
Results
A total of 643 patients of aged range 23–81 years (mean ± SD = 42.4 ± 13 years) were examined during the study. Among the 643 evaluated patients, 150 patients (23.3%) were suspected and positively diagnosed with various kinds of surgical site wound infection, as shown in Table 1. The significant factors for SSI development were age, previous antibiotic exposure, and type of exposure, while sex and antibiotic prophylaxis were not significantly associated with the development of differences, as shown in Table 2.
The surgical procedures of 150 postoperative surgical site infections were observed from patients who had laparotomy 63 (42%, n=150), appendectomy 42 (28%, n=150), prostatectomy 12 (8%, n=150), thyroidectomy 8 (5.3%, n=150), and others 25 (16.7%, n=150). The majority of the wound swabs, 122 (81.3%, n=150), developed bacterial growth within 18–24 h of incubation. A mixed growth pattern was observed in 41 patients out of 122 (33.6%, n=122), while a pure single bacterial growth was observed in 81 patients (66.4%, n=122). The rest, 28 patients (18.7%, n=150), did not observe any kind of bacterial growth even after incubation of 48 h and considered as the uninfected wound.
A total of 147 bacterial isolates were recovered in 122 patients where gram-positive bacteria were 44 (29.9%, n=147) and gram-negative bacteria were 103 (70.1%, n=147) (Table 3). Various bacterial pathogens with SSI, isolated from certain patients were Staphylococcus aureus, coagulase-negative Staphylococcus (CoNS), Enterococcus species, Klebsiella species, Escherichia coli, Pseudomonas aeruginosa, Enterobacter species, Citrobacter species, and Proteus species, were summarized (Table 4). The antimicrobial susceptibility patterns of gram-negative and gram-positive bacterial isolates are presented in Tables 5 and 6.
As shown in Table 4, Klebsiella spp. was considered as the first predominant gram-negative isolate, revealed a higher resistance level to ceftazidime and doxycycline, each 34 (89.4%, n=38); ampicillin 33 (86.8%, n=38); chloramphenicol 31 (81.5%, n=38); and gentamicin and ceftriaxone, each 27 (71%, n=38); and it was found to be sensitive to ciprofloxacin 25 (65.8%, n=38).
According to Table 4, the second predominant gram-negative isolate was E. coli, also exhibited the resistance against ampicillin and chloramphenicol, each 26 (83.8%, n=31); doxycycline 25 (80.6%, n=31); and gentamicin and ceftazidime, each 16 (51.6%, n=31). It was sensitive to ciprofloxacin 22 (71%, n=31). The first predominant gram-positive isolate, S. aureus, displayed some level of resistance to ceftazidime 17 (68%, n=25); doxycycline and gentamicin, each 13 (52%, n=25); chloramphenicol 12 (48%, n=25); ampicillin 11 (44%, n=25); ciprofloxacin and methicillin, each 9 (36%, n=25); and ceftriaxone 7 (28%, n=25), but all the S. aureus isolates exhibited sensitivity against vancomycin 25 (100%, n=25).
Discussion
Nosocomial infections become prominent in surgical wards because of surgical intervention and operative procedures. A total of 147 isolates were recovered from 122 patients in the present study over one and a half years.
In this study, the prevalence of SSI was 23.3%, in contrast to previous studies in Addis Ababa 39% [14], Mekelle Hospital 44% [10] in Ethiopia, and 7.3% in Pakistan [15]. The difference in surgical site infection’s magnitude may be due to the type of procedures, surgical setup, and environmental factors. Bacterial isolates were examined for the determination of its categories and antibacterial sensitivity patterns. The present analysis shows that gram-negative bacterial isolates predominate in SSIs; Klebsiella species (26%) is perhaps the most common isolated pathogen followed by Escherichia coli (21%), S. aureus (17%), P. aeruginosa (12%), CoNS (10%), Proteus species (4%), and Enterobacter species (4%). The current findings of the SSI infection-causing pathogenic pattern are different with earlier research in Bahir Dar [8] and Gondar [9, 16], where S. aureus is recorded as the widespread SSI-causing organism [17,18,19]. The reason for the predominancy of gram-negative bacteria might be because most of the infected patients are associated with abdominal surgery, and it is previously reported that gram-negative bacteria are predominantly related to intraabdominal procedures [20]. Findings of this research also indicated that more than 66% of gram-negative rods were resistant to ampicillin. A previous study also indicated that, in Gondar, Ethiopia, ciprofloxacin was effective for more than 90% of gram-negative isolates [9]. However, in the present study, ciprofloxacin was effective for more than 61% of the isolates. This sharp fall in effectiveness may be due to overuse of it as an empiric treatment option for most patients.
Among gram-negative isolates, Klebsiella species, Pseudomonas aeruginosa, and Escherichia coli demonstrated high resistance to most of the antibiotics tested. Eighty-four percent of Klebsiella species, 77% of Pseudomonas aeruginosa, and 61% of Escherichia coli were resistant to multiple antibiotics. Although they are not dependable for empiric treatment, ciprofloxacin and ceftriaxone were relatively useful for most bacterial isolates. This finding was also in agreement with the findings of other studies from Bahir Dar and Gondar, Ethiopia [8, 9, 16].
Resistance towards gentamicin in S. aureus differs globally; in our analysis, moderate resistance (52%) was observed that was greater than just that recorded at the university hospital in Gondar [16]. A higher incidence of methicillin resistance in S. aureus was also recorded in the present study; 36% of S. aureus isolates were methicillin-resistant Staphylococcus aureus (MRSA). The finding was higher than the recent report from Gondar University Hospital within the country (24%) [16], and in Mwanza, Tanzania (18.8%) [18].
In our experiment, gram-negative microbes exhibited higher resistance to widely recommended cost-effective antibiotics like penicillin derivatives, doxycycline, and chloramphenicol. The observed high resistance rate could be attributed because these low-priced antimicrobial agents are simple to administer, relatively inexpensive, and commonly prescribed in the evidential management of different infectious diseases. In order to efficiently treat surgical site infections, the use of such medications must be strictly examined for the therapeutic outcome and must be driven by microbiological tests. In the present study, resistance to the third-generation cephalosporins in gram-negative pathogens was higher, and these findings are similar to many prior reports from the Bahir Dar and Gondar regions of Ethiopia, and other nations [9, 16, 21]. An intensified unnecessary prescribing of ceftriaxone as a surgical prophylactic antibiotic at this referral hospital may be a potential cause for the observed greater resistance. Research on antibiotic usage in the same hospital reported ceftriaxone as the most frequently used injectable medication, with some more than 300 prescriptions each month in the surgical ward itself, as a piece of unpublished information from the hospital drugstore.
Besides, several patients from this study got ceftriaxone as a prophylactic antibiotic to avoid infections at the surgical site, and that may have disrupted the exposure of gram-negative pathogens susceptible to ceftriaxone. Most of the Pseudomonas aeruginosa strains isolated were highly sensitive to ceftazidime and moderately to ciprofloxacin and gentamicin. These findings agree with those studies from Gondar University Hospital [9, 16] and Karachi, Pakistan [15]. The above information demonstrates the ability of these classes of medicines for prescribing to treat Pseudomonas in SSIs.
Limitation of the study
Due to the limited resources, individual characterization of the isolates at the species level like Klebsiella spp., Proteus spp., Enterobacter spp., CoNS, Citrobacter spp., Enterobacter spp. was not possible to be performed.
Conclusions
The SSI rate was found to be 23.3%. The predominant isolates were Klebsiella spp, E. coli, S. aureus, P. aeruginosa, ConS, Proteus spp, and Enterobacter spp. The findings of research explored that gram-negative wound pathogens exhibited a significantly higher isolation rate (P<0.5) than gram-positive pathogens, with high resistance rates. Most gram-negative wound isolates were found to be resistant against commonly advised antibiotics. On the other hand, most isolated gram-positive pathogens were sensitive towards vancomycin, except one Enterococcus species. To achieve effective therapy for wound infections and to reduce/stop the appearance of multidrug-resistant (MDR) pathogens, continuous monitoring is essential with fair use of antimicrobial agents.
Availability of data and materials
The datasets used and analyzed during the study are available on reasonable request.
Abbreviations
- SSI:
-
Surgical site infection
- S. aureus :
-
Staphylococcus aureus
- E. coli :
-
Escherichia coli
- P. aeruginosa :
-
Pseudomonas aeruginosa
- CDC:
-
Centers for Disease Control and Prevention
- spp:
-
Species
- SOP:
-
Standard operating procedure
- SD:
-
Standard deviation
- CoNS:
-
Coagulase-negative Staphylococcus
- GN:
-
Gentamicin
- CIP:
-
Ciprofloxacin
- CEFTA:
-
Ceftazidime
- AMP:
-
Ampicillin
- CEF:
-
Ceftriaxone
- VAN:
-
Vancomycin
- CAF:
-
Chloramphenicol
- Met:
-
Methicillin
- DOX:
-
Doxycycline
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- MDR:
-
Multidrug resistant
References
Rhomberg PR, Fritsche TR, Sader HS, Jones RN (2006) Antimicrobial susceptibility pattern comparisons among intensive care unit and general ward gram-negative isolates from the Meropenem Yearly Susceptibility Test Information Collection Program (USA). Diagn Microbiol Infect Dis 56(1):57–62. https://doi.org/10.1016/j.diagmicrobio.2005.12.009
Weigelt JA, Lipsky BA, Tabak YP, Derby KG, Kim M, Gupta V (2010) Surgical site infections: causative pathogens and associated outcomes. Am J Infect Control 38(2):112–120. https://doi.org/10.1016/j.ajic.2009.06.010
Tacconelli E, De Angelis G, Cataldo MA, Mantengoli E, Spanu T, Pan A, Corti G, Radice A, Stolzuoli L, Antinori S, Paradisi F, Carosi G, Bernabei R, Antonelli M, Fadda G, Rossolini GM, Cauda R (2009) Antibiotic usage and risk of colonization and infection with antibiotic-resistant bacteria: a hospital population-based study. Antimicrob Agents Chemother 53(10):4264–4269. https://doi.org/10.1128/AAC.00431-09
Bibi S, Channa GA, Siddiqui TR, Ahmed W (2012) Pattern of bacterial pathogens in postoperative wounds and their sensitivity patterns. J Surg Pak (Int) 17(4):164–167
Al-Momany NH, Al-Bakri AG, Makahleh ZM, Wazaify MM (2009) Adherence to international antimicrobial prophylaxis guidelines in cardiac surgery: a Jordanian study demonstrates need for quality improvement. J Manag Care Pharm 15(3):262–271. https://doi.org/10.18553/jmcp.2009.15.3.262
Kotisso B, Aseffa A (1998) Surgical wound infection in a teaching hospital in Ethiopia. East Afr Med J 75(7):402–405
Tietjen L, Bossemeyer D, McIntosh N (2003) Infection prevention guidelines for healthcare facilities with limited resources. JHPIEGO Corporation, Baltimore, p 23–3
Fantahun B, Beyeh A, Atenaf A, Belay A (2009) Bacterial isolates from wound infection and their antimicrobial susceptibility pattern in Felege Hiwot referral hospital, Northwest Ethiopia. Ethiop J Health Sci 19(3):173–177
Mulu A, Moges F, Tessema B, Kassu A (2006) Pattern and multiple drug resistance of bacterial pathogens isolated from wound infection at University of Gondar Teaching Hospital, Northwest Ethiopia. Ethiop Med J 44(2):125–131
Tesfahunegn Z, Asrat D, Woldeamanuel Y, Estifanos K (2009) Bacteriology of surgical site and catheter related urinary tract infections among patients admitted in Mekelle Hospital, Mekelle, Tigray, Ethiopia. Ethiop Med J 47(2):117–127
Horan TC, Gaynes RP, Martone WJ, Jarvis WR, Emori TG (1992) CDC definitions of nosocomial surgical site infections, 1992: a modification of CDC definitions of surgical wound infections. Infect Control Hosp Epidemiol 13(10):606–608. https://doi.org/10.2307/30148464
Sangrasi AK, Leghari AA, Memon A, Talpur AK, Qureshi GA, Memon JM (2008) Surgical site infection rate and associated risk factors in elective general surgery at a public sector medical university in Pakistan. Int Wound J 5(1):74–78. https://doi.org/10.1111/j.1742-481X.2007.00365.x
College of Physicians & Surgeons of Saskatchewan Laboratory Quality Assurance Program (2010) Procedures/guidelines for the microbiology laboratory. College of Physicians & Surgeons of Saskatchewan Laboratory Quality Assurance Program, Saskatchewan
Endalafer N, Gebre-Selassie S, Kotiso B (2011) Nosocomial bacterial infections in a tertiary hospital in Ethiopia. J Infect Prev 12(1):38–43. https://doi.org/10.1177/1757177410376680
Mahmood A (2000) Bacteriology of surgical site infections and antibiotic susceptibility pattern of the isolates at a tertiary care hospital in Karachi. J Pak Med Assoc 50(8):256–259
Aschalew G, Solomon GS, Moges T, Eshetu M, Sisay Y (2014) Isolation of bacterial pathogens from patients with postoperative surgical site infections and possible sources of infections at University of Gondar Hospital, Northwest Ethiopia. J Environ Occup Health 3(2):103–108. https://doi.org/10.5455/jeos.20140512124135
Andhoga J, Macharia AG, Maikuma IR, Wanyonyi ZS, Ayumba BR, Kakai R (2002) Aerobic pathogenic bacteria in postoperative wounds at Moi Teaching and Referral Hospital. East Afr Med J 79(12):640–644. https://doi.org/10.4314/eamj.v79i12.8671
Mawalla B, Mshana SE, Chalya PL, Imirzalioglu C, Mahalu W (2011) Predictors of surgical site infections among patients undergoing major surgery at Bugando Medical Centre in Northwestern Tanzania. BMC Surg 11:21. https://doi.org/10.1186/1471-2482-11-21
Como-Sabetti KJ, Harriman KH, Fridkin SK, Jawahir SL, Lynfield R (2010) Risk factors for community-associated Staphylococcus aureus infections: results from parallel studies including methicillin-resistant and methicillin-sensitive S. aureus compared to uninfected controls. Epidemiol Infect 139(3):419–429. https://doi.org/10.1017/S0950268810001111
Koigi-Kamau R, Kabare LW, Wanyoike-Gichuhi J (2005) Incidence of wound infection after caesarean delivery in a district hospital in central Kenya. East Afr Med J 82(7):357–361
Anguzu JR, Olila D (2007) Drug sensitivity patterns of bacterial isolates from septic postoperative wounds in a regional referral hospital in Uganda. Afr Health Sci 7(3):148–154. https://doi.org/10.5555/afhs.2007.7.3.148
Acknowledgements
The authors would like to acknowledge the Arsi University Research and Publication Office for funding this project. The authors also thank the study participants, research and community service coordinators of the college of health sciences, Arsi University, and the microbiology laboratory technologists for collecting the sample and using the laboratory services. The authors are very thankful to the Asella Referral and Teaching Hospital surgical ward staffs for their cooperation during the data collection.
Funding
Arsi University Research and Publication Office, Asella, Ethiopia, financially supported our study. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
TG conceived and designed the research idea and performed proposal writing, data collection, laboratory work, and analysis. BB participated in the study design, analysis, and interpretations of the findings. MY participated in data analysis and interpretations, and write-up. SM participated in the study design, data collection, and laboratory work, and KMN participated in the data analysis and interpretations of the findings, write-up, manuscript writing, and reviewing. All authors were involved in reviewing the manuscript and approval for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was obtained from the College of Health Sciences Arsi University Ethical Review Committee (CHSAUERC), Arsi University, Asella, Ethiopia (approval reference number: A/CHS/RC005/15/16). The study was carried out at Asella Referral and Teaching Hospital, a public sector referral and teaching hospital affiliated to Arsi University College of Health Sciences. Written informed consent was obtained from each study participant. All the data collected from participant in the study was kept confidential.
Consent for publication
A consent for publication was obtained from all the participants while obtaining the consent to participate.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gebissa, T., Bude, B., Yasir, M. et al. Bacterial isolates and their antibiotic sensitivity pattern of surgical site infections among the surgical ward patients of Asella Referral and Teaching Hospital. Futur J Pharm Sci 7, 100 (2021). https://doi.org/10.1186/s43094-021-00255-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43094-021-00255-x