Abstract
Background
Growth performance, which can be evaluated using various body measurements, is crucial for providing red meat for human consumption. This study aims to improve the growth performance of sheep by identifying the different genotypes of leptin and IGFI genes associated with higher body measurements.
Methods
In this study, we collected blood samples from Barki (n = 30) and Farafra (n = 30) sheep, and measured their body height, width, length, and weight, and tail width and length to assess their growth performance. We used single strand conformation polymorphism polymerase chain reaction (SSCP-PCR) and sequencing analysis to identify single nucleotide polymorphisms (SNPs) and different genotypes of the leptin and insulin-like growth factor receptor (IGFI) genes associated with sheep body measurements.
Results
The leptin gene (209 bp) and IGFR gene (345 bp) were found to have three (TT, TG, and GG) and (TT, TC, and CC) genotypes, respectively, by SSCP-PCR. Sequence analysis revealed a SNP in the leptin gene (200G > T) with two alleles (G allele: OR058867 and T allele: OR058868). This SNP resulted in the substitution of the amino acid valine with leucine, which increased the width, length, and weight of the body and tail width in the heterozygous form (TG). The IGFI gene had three SNPs: 305 G > T in TT genotype (OR260670), 184C > T, 265T > C, and 305G > T in TC genotype (OR260669), and 265T > C in CC genotype (OR260671). The TC genotype of the IGFR gene was correlated with higher values of body measurements. Barki sheep were found to have the highest values for body weight, width, length, and tail width compared to Farafra sheep.
Conclusion
We recommend the use of entire Barki sheep in inbreeding programs to improve growth performance in Egyptian sheep.
Similar content being viewed by others
1 Background
Sheep are a vital source of meat and its by-products in dry areas. They provide a significant income for farmers and contribute 6.4% to the total red meat production in the Egyptian economy [1]. The most prevalent domestic breeds in Egypt include Al-Osaimi, Farafra, Al-Barky, and Al-Rahmani [2]. Barki sheep, raised under a transhumant system, graze on vegetation available from August to June and are well-suited to the harsh desert environment, including feed scarcity and high temperatures. Despite these conditions, they produce substantial amounts of meat, wool, and milk [3]. Farafra sheep, a native fat-tailed breed, are prevalent in the El-Farafra Oasis in the Egyptian western desert and have been kept under intensive management at the Mallawi Animal Production Research Station since 1992 [4]. Body weight and size can serve as indicators for future breeding and reproductive success in animals [5]. Several factors contribute to the significant variation in these measurements among sheep herds, including genetics, sex, birth type, and environmental conditions [6]. Recently, researchers have focused on specific body measurements that indicate an animal is well-fed and has a high meat yield, due to their economic importance [7]. Researchers have used molecular markers to study the association between candidate genes and performance traits in various sheep breeds, including milk traits in Sfakia sheep [8], carcass traits in Iranian Mehraban sheep [9], and tail types in Hu, Tibetan, and Dorper-Hu hybrid sheep [10]. Key candidate genes associated with milk and growth traits include PRL, LEP, IGF1, DGAT1, STAT5, CSN1S2, GHR, and GHRHR [11, 12].
The LEP gene, discovered in 1994 by Zhang [13], produces a protein composing 167 amino acids. This protein plays a role in growth, energy reserves, metabolism, immunity, reproduction, and regulating feed intake and body weight. It is a strong candidate gene for economically important traits in livestock [14]. Leptin plays a crucial role in regulating appetite and can affect factors related to eating, such as body weight and fat storage. Variations in the LEP gene have been linked to growth, body measurements, and carcass characteristics in various sheep breeds [15]. Polymorphisms in the LEP gene have also been shown to have additive effects on the weights and yields of primal cuts and carcasses [16].
IGF1 mediates many biological effects, including glucose absorption, myogenesis stimulation, apoptosis inhibition, cell cycle gene activation, lipid synthesis, progesterone production in granular cells, and DNA, protein, RNA synthesis and cell proliferation [17]. IGF1, a member of the IGF family (GHRH, GH, GHRHR, GHR, IGF-IR and IGF-IIR), is involved in processes such as reproduction, embryonic development, metabolism, cell differentiation, protein synthesis, skeletal growth, and fetal growth [18], also it was considered as a functional gene for a number of production traits such as meat and carcass quality traits, and growth in livestock. Variations in the IGF1 gene have been linked to growth traits in sheep [19]. Specific variants of the IGF1 gene have been associated with internal carcass length, rib yield, and neck weight in Santa Inês sheep. Additionally, a variant in the IGFI gene has been linked to the weights and yields of primal cuts such as rib, loin, leg, and neck [20]. It has been shown that IGFI gene polymorphisms had significant effects on many growth traits in Makooei and Balushi sheep [21, 22]. Hajhusseinlou et al. [23] studied the relationship between IGFI gene SNPs and several body traits in sheep. To our knowledge, this is the first study focusing on genotypes of leptin and IGFI genes associated with body measurements in Barki and Farafra sheep. This study examines mutations of the leptin and IGFI genes and their association with body measurements in Barki and Farafra sheep using SSCP-PCR, sequencing analysis, and appropriate statistical analysis.
2 Methods
2.1 Animal and blood samples
For the association analysis, 60 sheep were used as the experimental population, including 30 Barki and 30 Farafra sheep from the same farm. Blood samples, approximately 6 ml from vein of each animal, were taken by sterilized syringe and collected in tube containing EDITA. All samples were transferred into lab using icebox and stored at − 20 °C until DNA extraction.
2.2 Body measurements
A calibrated electronic scale was used to measure the body of Barki and Farafra sheep. The same persone used a clearly graduated leather ruler to measure the height and length of body as well as the length and width of tail. The all measurements were recorded and statistically analysed.
2.3 DNA extraction and PCR conditions
We used the Genomic DNA Kit (Geneaid, cat. No. GB 100) to extract DNA from whole blood samples, following the manufacturer’s instructions. We then measured the quantity and quality of the DNA using a NanoDrop 2000 spectrophotometer (Thermo, Waltham, MA, USA).
For the PCR reaction, we mixed 12.5 µL Master Mix, 1 µL of Primer-F, 1 µL of Primer-R, 4 µL of DNA, and 6.5 µL of ddH2O in a 25 µL reaction mixture. The primer sequence and PCR reaction were summarized in Table 1. We examined the PCR products on a 1% agarose gel.
2.4 Genotyping of leptin and IGFI genes and SNPs detection
We used the SSCP-PCR technique to study the genetic polymorphism of leptin and IGFI genes based on previous study for Darwish et al. [24]. a 5 μL of each PCR product was mixed with 45 μL of loading dye (98% Formamide, 10 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene-cyanol). After denaturation at 95 °C for 10 min, the samples were cooled on ice and loaded on 29% acrylamide:bisacrylamide gels. The gels were stained in a solution of 10% ethanol, 0.5% acetic acid, and 0.2% ethidium bromide for 10 min. Finally, the gels were washed with distilled water, and then photographed and analysed by gel documentation. Macrogen Co., Ltd. (Soul, Korea) sequenced the PCR products. We then compared the sequences using DNAMAN 6.0 software (https://www.lynnon.com) and Chromas 2 software (http://technelysium.com.au/wp/chromas) to identify any potential SNPs. These SNPs had been recorded at GenBank.
2.5 Statistical analysis
We calculated allele frequencies, polymorphic information content (PIC), heterozygosity (He), and the effective number of alleles (Ne) using Gene Calc, Copyright © 2023 according to Szymon and Jan [25]. We tested for Hardy–Weinberg equilibrium at each site using a chi-square test. We also used SPSS software version 18 to analyze the association between SNP genotypes and/or haplotypes and body measurements, using the following animal model:
In this model:
yi represents the trait measured in an individual. µ is the overall mean. Gi is a fixed effect corresponding to the genotype of polymorphisms. bi is the breed effect. ei is a random residual effect.
3 Results
3.1 Detection of leptin and IGFI genes and genotypes
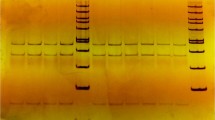
PCR detected the Leptin and Insulin-like Growth Factor 1 (IGF1) genes at 290 bp and 345 bp, respectively (Fig. 1a and b). SSCP-PCR revealed three genotypes for each gene. In Barki sheep, the leptin gene had TT (frequency 6), TG (frequency 17), and GG (frequency 7) genotypes. In Farafra sheep, the frequencies were 3, 11, and 16, respectively (Fig. 1c). The IGF1 gene had TT (frequency 19), TC (frequency 2), and CC (frequency 9) genotypes in Barki sheep. In Farafra sheep, the frequencies were 19, 4, and 7, respectively (Fig. 1d).
3.2 SNP Detection
Sequencing results revealed one SNP (200G > T) in the leptin gene with two alleles (T and G) as shown in Fig. 2. This SNP was genotyped and classified into TT, TG, and GG genotypes in an experimental population of 30 Barki sheep and 30 Farafra sheep (Fig. 3). The IGFI gene had three SNPs (184 C > T, 265 T > C, and 305 G > T) which were genotyped and classified into three genotypes in the same experimental population. These SNPs were recorded at GenBank under accession numbers: OR058867 for G allele and OR058868 for T allele of leptin gene. While IGFI gene had the following accession numbers: OR260670 for TT genotype, OR260669 for TC genotype, and OR260671 for CC genotype (Fig. 3a, b, and c).
a show SNPs of IGFI gene at position 184 C > T in TT genotype, b show SNP of IGFI gene at position 305C > T in TC, and c show SNP of IGFI at position 265T > C in CC genotype by sequence analysis, and d show alignment of IGFI gene fragment with sequence of IGFI deposited in gene bank under accession number X69474.1 by Bioedite software
3.3 Amino acids changes
In the leptin gene, a point mutation of the substitution type occurred at base 198G > T, resulting in a change of the amino acid valine to leucine, as shown in Fig. 4. All mutations (186 C > T, 266 T > C, and 306 G > T) in the IGFI gene occurred in the intron region.
3.4 Genetic parameters calculation
Table 2 displays the results of the genetic parameters calculation. All leptin gene sites were in Hardy–Weinberg equilibrium (p > 0.05). The OR058868 G > T site had moderate PIC status (0.25 < PIC < 0.5) in both Barki and Farafra sheep, with He and Ne values of 0.42–0.48 and 1.72–1.96, respectively. This site was highly variable and its alleles were evenly distributed. Similarly, all IGFI gene sites were in Hardy–Weinberg equilibrium (p < 0.05). The OR260670 C > T site had moderate PIC status (0.25 < PIC < 0.5) in both Barki and Farafra sheep, with He values of 0.42–0.46 and Ne values of 1.81 in Barki sheep and 1.96 in Farafra sheep. This site was also highly variable and evenly distributed, as shown in Table 3.
3.5 Effect of leptin genotypes, breed type, and interaction between breed and genotypes on body measurements
Barki sheep had better body measurements than Farafra sheep, with significantly higher body width (18.2 ± 0.6), tail width (15.1 ± 0.5), and tail length (29.5 ± 0.5) compared to Farafra sheep (8.9 ± 0.7, 8.7 ± 0.6, and 25.6 ± 0.6), respectively. However, animal height was significantly higher in Farafra sheep compared to Barki sheep (Table 4). Regarding the interaction between leptin genotypes and breed type, the TG genotype in Barki sheep had the highest significant value for animal height and body length traits, while no significant differences were observed in other traits between Barki and Farafra sheep (Table 4). The animal height trait was correlated with the GG genotype while body width, body length, tail width, and body weight traits were correlated with the TG genotype (Table 6).
3.6 Effect of IGFR genotypes, breed type, and interaction between breed and genotypes on body measurements
Body width, body length, and tail width traits were significantly higher in Barki sheep compared to Farafra sheep. Tail length and body weight traits were also better in Barki sheep than Farafra sheep. However, animal height was highest in Farafra sheep compared to Barki sheep (Table 5). Regarding the interaction between IGFI genotypes and breed type, the TC genotype in Barki sheep had the highest significant value for body width, body length, and tail width traits while the animal height trait was correlated with the TT genotype. No significant differences were observed in other traits between Barki and Farafra sheep (Table 5). Sheep with the TC genotype had significantly taller body length than TT and CC genotype carriers. The highest values of animal height, body width, tail width, and body weight were also correlated with the TC genotype (Table 6).
4 Discussion
The effectiveness of using markers to select desirable traits in animals for sale relies on several factors. These include finding markers that are closely linked to the genes responsible for the traits, verifying that variations in these genes are present in the population, and understanding how these genes interact with other genes or environmental factors that affect economically important traits. This must be done in a cost-effective and efficient manner for the technology to be adopted by livestock industries [26]. These genes can be closely linked to genetic markers and can be inherited together. A good genetic marker should have multiple forms, be inherited in a simple manner, and be easy to detect [27]. There are two ways to study the genes responsible for variations in phenotypic characteristics. The first is to start with the phenotypic characteristic and work toward the genome, using association mapping based on linkage disequilibrium or by targeting specific candidate genes based on their similarity to known genes. The second is to start with the genome and work toward the phenotypic characteristic, using statistical analysis of genomic data to identify likely targets of past selection through selective sweep analysis [28, 29].
Fat tail breeds make up approximately 25% of the world’s sheep population and are grazed in many countries, particularly in Asia and northern Africa. Sheep breeding plays a vital role as a source of protein through increasing body weight of sheep, thus improving the local economy in these countries. The variants of genes associated with fat tail trait affecting the phenotypic expression are of both theoretical and economic interest. The fat tail is an important trait for coping with environmental hazards and serves as a valuable energy reserve for animals during migration and winter. However, the desire of producers and consumers for fat-tailed sheep decreased in the intensive and non-intensive systems. As a result in the industrialized country, fat tail size reduction could be a goal in sheep. In sheep breeding, tail type traits have a significant impact on performance [11]. Due to the presence of fat in the tail, it has additional value to humans because it provides high-energy food during times of drought and famine. Nowadays, the requirements for high-quality meat consumption have increased, and the basic points for determining meat quality are the distribution and fat content of the animal carcass [30]. Behinds, the fats found in sheep tails are considered healthy for humans as they are unsaturated fats and they also reduce cooking time [31], and the current study showed that there is a positive relationship between fat tail and high body weight, which may change this tendency for producers and consumers. In many sheep studies, including those on growth, inheritance, and nutrition, measurements of size and body shape are important [11]. For species raised for meat, body shape and growth rate are important selection criteria. The average weight of the fat tail was 1.03 kg, and the ratio of tail fat to carcass varied widely [32]. This could be an indicator of biological diversity in this breed and could be used to select lambs with smaller fat tails. Compared to Turkish Awassi sheep, where the average tail fat weight was 3 kg and accounted for 15% of the carcass weight [33], as well as compared to the Lori, Mehraban, and Ghezel breeds of Iran [34], the fat tail of the Makoei breed was relatively light.
Indeed, information on the relationship between genetic polymorphisms and tail fat measurements in sheep is scarce. Most investigations have focused on the correlation of carcass traits with genetic polymorphisms of related genes. In fact, there are few reports were done on association of fat tail traits with presence of SNPs in the IGF-1 and PIT-1 genes of sheep. Recently, few polymorphisms of the IGF-1 and PIT-1 genes have been detected in sheep compared to other small ruminants. Little information is available to compare the different Iranian sheep breeds. This is the first study that attempts to reveal the relationship between allele variation in sheep IGF-1 and leptin genes and body measurements associated with tail traits in Egyptian sheep. The current study found that the TC genotype of the IGFI gene in Barki sheep had the highest significant value for body width, body length, and tail width traits while the animal height trait was correlated with the TT genotype. Sheep with the TC genotype of the IGFI gene had significantly taller body length than TT and CC genotype carriers. The highest values of animal height, body width, tail width, and body weight were also correlated with the TC genotype. There significant correlations were found for all fat tail measurement traits with genotypes of the IGF-1 and PIT-1 genes in the Makooei sheep. Sheep with longer, wider tails have the P3 genotype of the PIT-1 gene while sheep with thicker tails have the P4 genotype [35]. In Lori Bakhtiari and Zil breeds, the exon 17 region of the DGAT1 gene was associated with carcass weight and dressing percentage in sheep [36]. A significant association was also found in Makooei sheep between various transferrin proteins genotypes and fat tail weight, hot carcass weight, and hot carcass weight free of fat tail [37]. A nearly significant relationship was reported between band patterns and tail length and tail down circumference in the fourth exon of the growth hormone gene in Kermanian sheep [12]. Yakar et al. [38] showed that IGF-1 plays an important role in skeletal development, while Sun et al. [39] found a connection between IGF1 gene expression and body weight in Hu sheep. The IGF1 gene is crucial for animal growth, including cell proliferation, maintenance of skeletal muscle satellite cells for regeneration, and muscle hypertrophy [40]. An association between BL, TW and body weight traits in Balami sheep and WH traits in Yankasa sheep with IGF1 gene was detected by PCR–RFLP [41]. A PCR-SSCP in exon 1 was linked to ADG and BL in Makui sheep [42], and ADG in both Baluchi [43] and Makooei [44] sheep breeds. The IGF-I AA genotype was found with a frequency of 83.3% in Colored Polish Merino sheep, had significant effects on meat color, outer carcass fat, and drip loss in the study by Grochowska et al. [45], and were associated with longissimus dorsi, width and flavor of muscle. Leptin helps regulate the balance between food intake and energy expenditure by sending signals to the hypothalamus [46]. Due to its ability to break down fat and regulate fat stores, genetic variations in LEP may be important in mobilizing lipids for milk production, with potential implications for the body weight of offspring in sheep. In line with this, genetic variations in leptin have been shown to affect milk performance in cattle [47]. The LEP gene is considered a candidate gene that affects body fat content [48]. A LEP polymorphism was also found to be significantly associated with milk yield in Najdi ewes from Saudi Arabia [49]. Several studies have highlighted the role of LEP in growth traits in sheep [50]. Statistical analysis results of recent study showed a significant relationship between Leptin gene (exon 3) and tail, chest, abdomen, and neck circumference as well as backfat thickness. This study revealed new associations where the C allele had a positive effect on fat tail weight and back fat thickness in fat-tailed sheep [51]. The SNP of leptin gene in position A113G was associated with an increase in fat-tail percent, carcass weight, and total body fat weight in the Shal sheep breed while it was associated with reduction in carcass weight, lean meat weight, and slaughter weight, and an increase in fat-tail percent [52]. The polymorphism of Leptin gene at position 332G > A had significant effect on body length. This non-synonymous mutation caused change in amino acid from Arg to Gln [15]. The present study found that the TG genotype of the Leptin gene in Barki sheep had the highest significant value for animal height and body length traits, while no significant differences were observed in other traits between Barki and Farafra sheep. The animal height trait was correlated with the GG genotype while body width, body length, tail width, and body weight traits were correlated with the TG genotype. There are haplotypes in IGF1 and LEP genes associated with body traits in Santa Inês sheep [53]. Therefore, this study concluded that SNPs in the leptin and IGFI genes could be useful for improving growth performance in sheep.
5 Conclusion
The present study found that the leptin and IGFI genes had these genotypes: TT, TG, and GG for leptin, and TT, TC, and CC for IGFI. Sequence analysis of the leptin gene revealed two alleles with one SNP, resulting in the substitution of valine with leucine. This increased body and tail width, length, and weight in the TG heterozygous form. The IGFI gene had three SNPs in intron 1 region, with the TC genotype correlating with higher body measurements. Barki sheep had higher values for body weight, width, length, and tail width compared to Farafra sheep. These results are important in breeding programs to improve body measurements in Egyptian sheep.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- SSCP-PCR:
-
Single strand conformation polymorphism polymerase chain reaction
- SNP:
-
Single nucleotide polymorphisms
References
Yagoubi Y, Smeti S, Ben Saïd S, Srihi H, Mekki I, Mahouachi M, Atti N (2021) Carcass traits and meat quality of fat-tailed lambs fed rosemary residues as a part of concentrate. Animals 11(3):655. https://doi.org/10.3390/ani11030655
Aboul-Naga AM, Alsamman AM, El Allali A, Elshafie MH, Abdelal ES et al (2022) Genome-wide analysis identified candidate variants and genes associated with heat stress adaptation in Egyptian sheep breeds Front. Genet 13:898522. https://doi.org/10.3389/fgene.2022.898522
Hekal SA, Badawy NS, Mona IM (2017) Skin and fleece characteristics of barki sheep as affected by docking. EJAP 54(3):193–198
Bashandy T, Hussein A, Solma M, Kassab A, Hamdon H (2020) Molecular evaluation of three populations of farafra sheep in comparison to Ossimi and Rahmani sheep breeds. Acta Univ Agric Silvic Mendel Brun. https://doi.org/10.11118/actaun202068060929
Tariq MM, Eyduran E, Bajwa MA, Waheed A, Iqbal F, Javed Y (2012) (2012) Prediction of body weight from testicular and morphological characteristics in indigenous Mengali sheep of Pakistan: using factor analysis scores in multiple linear regression analysis. Int J Agric Biol 14:590–594
Gurcan IS. Estimaion of live weight by statistical methods using body measurements in Merino sheep. Ank Üniv Health Sci Inst. PhD Thesis, Ankara. 2000
Al-Enzy AFM, Saed ZJM, Naser AS, Mohammed T et al (2020) The role of adding sodium chloride in broiler chicks diets to improve production performance and antioxidant status during heat stress. ATMPH 23(16):231–612. https://doi.org/10.36295/ASRO.2020.231612
Ibrahim AH, Tzanidakis N, Sotiraki S, Zhou H, Hickford JG (2019) Identification of the association between FABP4 gene polymorphisms and milk production traits<? xmltex\break?> in Sfakia sheep. Arch Anim Breed 62(2):413–422
Li S, Zhou H, Zhao F, Fang Q, Wang J, Liu X, Luo Y, Hickford JGH (2021) Nucleotide sequence variation in the insulin-like growth factor 1 gene affects growth and carcass traits in New Zealand Romney Sheep. DNA Cell Biol 40(2):265–271. https://doi.org/10.1089/dna.2020.6166
Li Q, Lu Z, Jin M, Fei X, Quan K, Liu Y, Ma L, Chu M, Wang H, Wei C (2020) Verification and analysis of sheep tail type-associated PDGF-D gene polymorphisms. Animals 10(1):89. https://doi.org/10.3390/ani10010089
Gavran M, Antunović Z, Gantner V (2021) Candidate genes associated with economically important traits of sheep-a review. Agric Conspec Sci 86(3):195–201
Hajihosseinlo A, Jafari S, Ajdary M (2015) The relationship of GH and LEP gene polymorphisms with fat-tail measurements (fat-tail dimensions) in fat-tailed Makooei breed of Iranian sheep. Adv Biomed Res 4:172. https://doi.org/10.4103/2277-9175.163995
Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM (1994) Positional cloning of the mouse obese gene and its human homologue. Nature 372:425–432. https://doi.org/10.1038/372425a0
Caron A, Lee S, Elmquist JK, Gautron L (2018) Leptin and brain-adipose crosstalks. Nat Rev Neurosci 19:153–165. https://doi.org/10.1038/nrn.2018.7
Bakhtiar R, Abdolmohammadi A, Hajarian H, Nikousefat Z, Kalantar-Neyestanaki D (2017) Identification of 332G > A polymorphism in exon 3 of the leptin gene and partially effects on body size and tail dimension in Sanjabi sheep. Int J Bioeng Life Sci 11:506–509
Meira AN, Moreira GCM, Coutinho LL, Mourão GB et al (2018) Carcass and commercial cut yield of Santa Inês sheep affected by polymorphisms of the LEP gene. Small Rumin Res 166:121–128. https://doi.org/10.1016/j.smallrumres.2018.06.012
Chelongar R, Hajihosseinlo A, Ajdary M (2014) The effect of Igf-1 and pit-1 genes polymorphisms on fat-tail measurements (fat-tail dimensions) in Makooei sheep. Adv Environ Biol 14:862–868
Masoud N, Abbas H, Marziyeh A (2013) PCR-SSCP variation of IGF1 and PIT1 genes and their association with estimated breeding values of growth traits in makooei sheep. Gene Res Int 272346:6. https://doi.org/10.1155/2013/272346
Kader Esen V, Esen S (2023) Association of the IGF1 5’UTR polymorphism in meat-type sheep breeds considering growth, body size, slaughter, and meat quality traits in Turkey. Vet sci 10(4):270. https://doi.org/10.3390/vetsci10040270
Meira AN, Montenegro H, Coutinho LL, Mourão GB et al (2019) Single nucleotide polymorphisms in the GH and IGF1 genes associated with carcass traits in Santa Inês sheep. Animal 13:460–468. https://doi.org/10.1017/S1751731118001362
Negahdary M, Hajihosseinlo A, Ajdary M (2013) PCR-SSCP vari ation of IGF1 and PIT1 genes and their association with esti mated breeding values of growth traits in Makooei Sheep. Genet Res Int 6:272346. https://doi.org/10.1155/2013/272346
Gholibeikifard A, Aminafshar M, Hosseinpour MM (2013) Polymorphism of IGF-I and ADRB3 genes and their association with growth traits in the iranian baluchi sheep. J Agr Sci Tech 15:1153–1162
Hajihosseinlo A, Hashemi A, Razavi-Sheshdeh SA, Pi rany N. (2013) Association of the polymorphism in the 50 flanking re gion of the ovine IGF-I gene with growth and development traits in Makui sheepof Iran. Eur J Zoolog Res 2:19–24
Darwish AM, Darwish HR, Mabrouk DM, Abdelhafez MA, Abdel-Salam AM, Mohamed IE, Farag IM (2022) Effectiveness of β-lactoglobulin and Leptin genes variants on zaraiby and damascus goat milk traits. Adv Anim Vet Sci 10(5):1113–1118. https://doi.org/10.17582/journal.aavs/2022/10.5.1113.1118
Szymon M, Jan B. Gene Calc, Copyright © 2023 https://gene-calc-.pl./pic
Moore SS, Li C, Basarab J, Snelling WM et al (2003) Fine mapping of quantitative trait loci and assessment of positional candidate genes for backfat on bovine chromosome 14 in a commercial line of Bos taurus. J Anim Sci 81:1919–1925. https://doi.org/10.2527/2003.8181919x
Thaller G, Kuhn C, Winter A et al (2003) DGAT1, a new positional and functional candidate gene for intramuscular fat deposition in cattle. Anim Genet 34:354–357. https://doi.org/10.1046/j.1365-2052.2003.01011.x
Akey JM (2009) Constructing genomic maps of positive selection in humans: where do we go from here. Genome Res 19(5):711–722. https://doi.org/10.1101/gr.086652.108
Duguma G, Cloete SWP, Schoeman SJ, Jordaan GF (2002) Genetic parameters of testicular measurements in Merino rams and the influence of scrotal circumference on total flock fertility. S Afr J Anim Sci 32(2):76–82. https://doi.org/10.4314/sajas.v32i2.3748
Moradi MH, Nejati-Javaremi A, Moradi-Shahrbabak M, Dodds KG, McEwan JC (2012) Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet 13(1):1–15
Öztürk K, Yılmaz OZF, Kaya M, Kaban G (2023) The effects of sheep tail fat, fat level, and cooking time on the formation of Nε-(carboxymethyl) lysine and volatile compounds in beef meatballs. Foods 12(15):2834
Mohapatra A, Shinde AK (2018) Fat-tailed sheep-an important sheep genetic resource for meat production in tropical countries: an overview. IJSR 24(1):1–17. https://doi.org/10.5958/0973-9718.2018.00020.X
Dekker JCM (2004) J Anim Sci 82:313–328
Sadeghi S, Hajihosseinlo A, Bohlouli M (2014) Haplotype association of ovine leptin gene on breeding value of body measurements in Makooei sheep breed. Biotechnol Anim Husb 30(2):233–242. https://doi.org/10.2298/BAH1402233S
Chelongar R, Abbas H, Marziyeh A. The effect of Igf-1 and pit-1 genes polymorphisms on fat-tail measurements (fat-tail dimensions) in Makooei sheep. Adv Environ Biol. 2014; 862–868.
Farahani AK, Shahrbabak HM, Shahrbabak MM, Yeganeh M (2010) Relationship of fat-tail and body measurements with some economic important traits in fat-tail Makoei breed of Iranian sheep. Afr J Biotechnol 9(36):5989–5992
Abousoliman I, Reyer H, Oster M, Muráni E, Mourad M, Abdel-Salam Rashed M, Mohamed I, Wimmers K (2020) Analysis of candidate genes for growth and milk performance traits in the Egyptian Barki sheep. Animals 10(2):197. https://doi.org/10.3390/ani10020197
Yakar S, Werner H, Rosen CJ (2018) 40 Years of IGF1 Insulin-like growth factors: actions on the skeleton. J Mol Endocrinol 61:T115–T137. https://doi.org/10.1530/JME-17-0298
Sun W, Su R, Li D, Musa HH et al (2014) Developmental changes in IGF-I and MyoG gene expression and their association with meat traits in sheep. GMR 13:2772–2783. https://doi.org/10.4238/2014.April.14.6
Philippou A, Maridaki M, Halapas A, Koutsilieris M (2007) The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In Vivo 21:45–54
Raji AO, Mohammed A, Igwebuike JU, Alphonsus C (2017) Association of IGF 1 gene polymorphisms with some morphometric traits of Nigerian indigenous sheep breeds. Niger J Biotechnol 34:97–104. https://doi.org/10.4314/njb.v34i1.13
Hajihosseinlo A, Hashemi A, Razavi-Sheshdeh SA, Pirany N (2013) Association of the polymorphism in the 5’ flanking region of the ovine IGF-I gene with growth and development traits in Makui sheep of Iran. Eur J Zool Res 2:19–24
Tahmoorespur M, Valeh MV, Nassiry MR, Moussavi AH, Ansary M (2009) Association of the polymorphism in the 5’ flanking region of the ovine IGF-I gene with growth traits in the Baluchi sheep. S Afr J Anim Sci 39:97–101. https://doi.org/10.4314/sajas.v39i1.61319
Negahdary M, Hajihosseinlo A, Ajdary M (2013) PCR-SSCP variation of IGF1 and PIT1 genes and their association with estimated breeding values of growth traits in Makooei sheep. Genet Res Int. https://doi.org/10.1155/2013/272346
Grochowska E, Borys B, Janiszewski P, Knapik J, Mroczkowski S (2017) Effect of the IGF-I gene polymorphism on growth, body size, carcass and meat quality traits in Colored Polish Merino sheep. Arch Anim Breed 60(2):161–173
Bhowmik N, Ringwall KA, Dahlen CR, Swanson KC, Clapper JA, Ward AK, Hulsman Hanna LL (2019) The role of leptin in reproductive characteristics of commercial beef cows and heifers. Transl Anim Sci 1:1764–1768. https://doi.org/10.1093/tas/txz083
Mahmoud A, Saleh A, Almealamah N et al (2014) Polymorphism of leptin gene and its association with milk traits in Najdi sheep. J Appl Microbiol 8:2953–2959
Zhou H, Hickford JGH, Gong H (2009) Identification of allelic polymorphism in the ovine leptin gene. Mol Biotechnol 41:22–25. https://doi.org/10.1007/s12033-008-9090-3
Yang D, Chen H, Wang X, Tian Z, Tang L, Zhang Z (2007) Association of polymorphisms of leptin gene with body weight and body sizes indexes in Chinese indigenous cattle. J Genet Genomics 34:400–405. https://doi.org/10.1016/S1673-8527(07)60043-5
Tahmoorespur M, Taheri A, Valeh MV, Saghi DA, Ansary M (2010) Assessment relationship between leptin and ghrelin genes polymorphisms and estimated breeding values (EBVs) of growth traits in Baluchi sheep. J Anim Vet Adv 9:2460–2465
Kader Esen V, Esen S (2023) Association of the IGF1 5′ UTR polymorphism in meat-type sheep breeds considering growth, body size, slaughter, and meat quality traits in Turkey. Vet Sci 10(4):270. https://doi.org/10.3390/vetsci10040270
Barzekar R., Aar S, Frouzandeh M. Polymorphisms of the ovine leptin gene and its association with growth and carcass traits in three Iranian sheep breeds. 2009; 241–246.
Machado AL, Meira AN, Jucá AF, Azevedo HC et al (2020) Variants in GH, IGF1, and LEP genes associated with body traits in Santa Inês sheep. Sci Agric. https://doi.org/10.1590/1678-992X-2019-0216
Acknowledgment
The authors acknowledge Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R5), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
Funding
This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R5), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
Author information
Authors and Affiliations
Contributions
This study was done in collaboration with all authors. AD designed this study. AD, MAA, and ZA participated in the conduct of the study. AD, AA and SO analyzed the data. AD drafted the manuscript. AD and ZA critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The experimental procedure used in this investigation was approved by the Animal Care and Use Committee of National Research Centre in Egypt
Consent for publication
Not applicable.
Competing interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Darwish, A.M., Abdelhafez, M.A., Abdel-Hamid, Z.G. et al. Correlation analysis between polymorphism of leptin and IGFI genes and body measurements in Barki and Farafra sheep. Beni-Suef Univ J Basic Appl Sci 12, 119 (2023). https://doi.org/10.1186/s43088-023-00450-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43088-023-00450-0