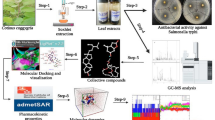
Abstract
Background
Salmonella belongs to the Enterobacteriaceae family, a gram-negative, non-spore-forming, rod-shaped, motile, and pathogenic bacteria that transmit through unhygienic conditions. It is estimated that 21 million new infections arise every year, resulting in approximately 200,000 deaths. It is more prevalent among children, the old aged, and immunocompromised individuals. The frequent usage of classical antimicrobials has begun the increasing emergence of various drug-resistant pathogenic bacterial strains. Hence, this study was intended to evaluate the bioactive seaweed sulfated polysaccharides (SSPs) against the ompF (outer membrane porin F) protein target of Salmonella typhi. SSP is the sulfated compound with a wide range of biological activities, such as anti-microbial, anti-allergy, anti-cancer, anti-coagulant, anti-inflammation, anti-oxidant, and anti-viral.
Results
In this study, eleven compounds were targeted against S. typhi OmpF by the molecular docking approach and were compared with two commercially available typhoid medications. The SSP showed good binding affinity compared to commercial drugs, particularly carrageenan/MIV-150, carrageenan lambda, fucoidan, and 3-phenyllactate, ranked as top antagonists against OmpF. Further, pharmacokinetics and toxicology (ADMET) studies corroborated that SSP possessed drug-likeness and highly progressed in all parameters.
Conclusions
AutoDockTools and Schrodinger's QikProp module results suggest that SSP could be a promising drug for extensively drug-resistant (XDR) S. typhi. To the best of our knowledge, this is the first report on in silico analysis of SSP against S. typhi OmpF, thus implying the capabilities of SSPs especially compounds like carrageenans, as a potential anti-microbial agent against Salmonella typhi infections. Eventually, advanced studies could corroborate SSPs to the next level of application in the crisis of XDR microbial diseases.
Graphical Abstract

Similar content being viewed by others
1 Background
Salmonella belongs to the Enterobacteriaceae family, a gram-negative, non-spore-forming, rod-shaped, motile, and pathogenic bacteria that transmit through unhygienic conditions. The molecular morphology of Salmonella serovars contains O, K, H, and the Vi capsular antigens (https://salmonellatyphi.org/). Among these serotypes, Salmonella enteritidis and Salmonella typhi have significant adverse effects and have been reported to cause severe and occasional life-threatening diseases like typhoid fever, sepsis, diarrhea, and gastroenteritis [12]. It is more prevalent among children, old aged, immunocompromised individuals, and it is estimated that 21 million new infections emerge every year, resulting in approximately 200,000 deaths [8]. They were primarily contributed by Southeast Asia, Southcentral Asia, Turkey, Morocco, Southern Africa, and Latin America. As there is a hike in travelers worldwide, it has been estimated that the incidence rate of typhoid fever in travelers to high-endemic countries is 3–30 cases per 100,000 travelers [22]. The frequent usage of classical antimicrobials has begun the increasing emergence of various drug-resistant pathogenic bacterial strains. An outbreak was reported from 2016 to 2018 in Pakistan, a large typhoid fever resulted in 5372 XDR (Extensively drug-resistance) typhoid cases transmitted across the country. This XDR strain is only susceptible to azithromycin and ciprofloxacin and was only preventable by improving the hygienic conditions, improved access to clean water, sanitation, and vaccination and anti-microbial therapy [6]. In the last three decades, research showed the emergence of multidrug-resistant (MDR) S. typhi, a huge threat to humankind. S. typhi has acquired resistance against chloramphenicol, ampicillin, trimethoprim/sulfamethoxazole, and they are susceptible to fluoroquinolones, cephalosporins (ceftriaxone), and azalide azithromycin. The phylogeographical analysis of the S. typhi shows that H58 lineage is highly associated with the MDR and reduced susceptibility to fluoroquinolones such as ciprofloxacin. As a result of MDR, there is an urge to discover new antibiotics or alternative therapeutics to treat S. typhi [26]. At an instant, seaweeds got the researcher's attention to treat infectious diseases because of their abundant availability in nature with major health benefits [19]. Seaweed sulfated polysaccharides (SSPs) are a complex group of macromolecules with a wide range of important biological activities, such as anti-microbial, anti-allergy, anti-cancer, anti-coagulant, anti-inflammation, anti-oxidant, and anti-viral properties. SSP is commonly found in all three major groups of seaweeds: brown, green, and red seaweeds. Brown SSPs are fucans, including fucoidan, sargassan, ascophyllan, and glucuronoxylofucan; red SSPs are galactans that are commercially available as agar and carrageenan. Green SSPs are usually available as sulfated heteropolysaccharides including galactose, xylose, arabinose, mannose, glucuronic acid, or glucose [27]. On the other hand, 3-phenyl lactic acid is also reported as a natural, effective anti-microbial drug against the pathogen Salmonella [28].
Molecular docking is a virtual screening tool applied in drug discovery with the best binding affinities to predict macromolecules (target protein) and small molecules (ligand) interactions that predict the molecules' conformation and orientation. Hence, this study was intended to evaluate the bioactive SSP against the Ompf (outer membrane porin F) protein target of Salmonella typhi. To the best of our knowledge, this is the first report on sulfated polysaccharides from seaweeds against S. typhi. In this present study, the molecular findings of the SSP as a natural antagonist against OmpF of S. typhi by the in silico approach with ADMET profiling are performed. This study would help to understand the mechanism of action at the molecular level. Eventually, advanced studies could corroborate SSPs to the next level of application in the crisis of XDR microbial diseases.
2 Methods
2.1 Target preparation
The outer membrane protein (OmpF) of Salmonella typhi was chosen as the target, and the structure was retrieved from the Protein Data Bank (PDB ID: 4KR4). The inhibitor and HETATM were removed by the Discovery Studio 2020 (Dassault System BIOVIA) visualization tool and saved as PDB format for further molecular docking studies.
2.2 Ligand preparation
SSPs were used as a possible anti-bacterial compound (ligand) against OmpF of S. typhi. The structure of bioactive SSP compounds and commercially available drugs for typhoid fever were retrieved from the PubChem Database (Table 1, Fig. 1) in the format of structure data file (SDF); energy minimization was done using Molecular Mechanics Force Field (MMFF) in PyRx virtual screening tool and saved as PDB format for further molecular docking studies.
2.3 Molecular docking studies
The computational studies were carried using AutoDockTools 1.5.6 (ADT) (Scripps Research US) with the extension suite to the Python Molecular Viewer of MGL tools with the Cygwin program. The protein and ligand were imported and preprocessed with the removal of water molecules, addition of polar hydrogens, and merging of nonpolar with Kollman and Gasteiger charges. Then, the grid parameter file and docking parameter file were prepared. The program was executed on the Cygwin platform using the Lamarckian genetic algorithm (LMA) with 50 independent runs [2]. The resulting ideal docking outputs were analyzed by the orientation and conformation with the least binding score as best affinity. Further, the molecular interaction and distance were calculated in Discovery Studio visualization tool.
2.4 In silico pharmacokinetic and toxicological profiling
The drug-likeness assessment based on adsorption, distribution, metabolism, excretion, and toxicity (ADMET) actions of SSP compounds in the biological network was computationally evaluated using the QikProp module on Schrodinger's Maestro platform. The pharmaceutically important properties of the substances were predicted, and the physicochemical descriptors were compared with the reference values of the QikProp user manual (v3.5, Schrödinger).
3 Results
The medication for bacterial diseases is becoming increasingly difficult because of the development of antimicrobial drug resistance, especially by gram-negative bacterial pathogens like Salmonella [17]. Extensively utilized antibacterial drugs act by hindering cell wall synthesis and binding with target proteins of bacteria. The outer membrane of gram-negative bacteria provides a protective barrier against toxic compounds and environmental stress. OmpF is an integral membrane protein that functions as a porin, involving in transmembrane ion, nutrient molecules, and waste product transport [7, 10]. Various reports have shown that in OmpF protein's destruction, mutation is associated with altered drug susceptibility in Salmonella typhimurium [4, 5, 16, 20]. As a result, a greater comprehension of how changes in membrane permeability activate bacterial resistance is required to develop further antibiotic treatment approaches [29].
3.1 Molecular docking analysis
OmpF porin is a trimeric integral membrane protein responsible for the passive transport of small hydrophilic molecules, such as nutrients and waste products, across the outer membrane. In this study, totally eleven compounds were subjected to the molecular docking study. Among them, eight compounds were SSP, one was 3-phenyllactate, and two commercial typhoid drugs, namely azithromycin and ciprofloxacin, were docked with OmpF of S. typhi. The best binding scores were obtained in Carrageenan/MIV-150 with a binding score of − 4.61 kcal/mol, followed by carrageenan lambda (− 4.95 kcal/mol); 3,6-anhydro-D-galactose-2-sulfate (− 3.26 kcal/mol) and 3-phenyllactate (− 3.59 kcal/mol). The hydrogen (H bond) interactions are described in Table 1. The two- and three-dimensional molecular interactions of protein–ligand complexes are depicted in Figures 2, 3, 4, and 5.
3.2 In silico pharmacokinetic and toxicological profiling
The investigation of drug passages across the anatomy by computational approach is a superficial tool to predict the pharmacology in a complicated biological system. The prediction of functionally indicative descriptions and pharmaceutically pertinent attributes of organic molecules can be meticulously estimated by the QikProp module. The assessment of pharmacokinetics of drug molecule essentially needs to be analyzed prior to implementation of clinical trial. Even highly bioactive compound often fails to other physical and metabolic factorials. In this work, we have analyzed the peculiarity nature of the nine SSP ligand molecules at various physical and biological criteria. Particularly the adsorption on Caco cell lines, oral adsorption, crossing over the blood–brain barrier, skin permeability, donor and acceptor of hydrogen bond, and Lipinski’s rules results are tabulated in Table 2. All the nine SSP ligand molecules exhibited significant rating within the specified limitation. Especially the polarizing nature of ligand molecules is an immense need to be dispersed over the blood stream. κ-Carrageenan exhibited 53.668 cubic angstroms of polarizability, whereas 3,6-anhydro-2-(hydrogen sulfate)-alpha-D-galactopyranose exhibited 14.888 cubic angstroms that is categorized under recommended values of the QikProp Manual. The molecular interaction of SSP ligand molecules was predicted by the sharing of donor and acceptor hydron bonds in complex biological systems wherein κ-carrageenan showed 10 and 36.9 range of donor and acceptor hydron bonds, respectively, which leads to functionally interactive configurations. In contrast, the output of Caco cell permeability of κ-carrageenan was lesser degree (0.006) that falls below the recommended values as shown in Table 2. The highly prominent gut cell (Caco) adsorption was appeared in β-carrageenan, which showed 312.18 in the recommended range of > 25 to < 500 values. The α-carrageenan, κ-carrageenan, carrageenan-lambda, and λ-carrageenan were ahead of the recommended value in the prediction of blood–brain barrier co-efficiency that reveals the sign of crossing over the blood–brain barrier. Finally, the determination of route of administration either oral or blood stream was resolved by the human oral adsorption features of drug molecules. Four out of nine SSP ligands such as carrageenan/MIV-150, β-carrageenan, fucoidan, and 3-phenyllactate showed better results in human oral adsorption. Other SSPs can be administrated through blood stream or improved medicinal formulation for greater oral adsorption. Interestingly, carrageenan/MIV-150, β-carrageenan, and fucoidan compounds passed the criteria and highly progressed in all parameters (superior range are bolded in Table 2). The results evidenced that the ligand molecules from seaweeds can be successfully employed against Salmonella typhi in human system without loss of its anti-microbial activity.
4 Discussion
SSP is an abundant molecule in marine algal system with extended pharmacological applications. There are limited studies reported for anti S. typhi activity from traditional medicinal plants like Vitex doniana (root), Cassia tora (Leaf), Alstonia boonei (bark), Stachytarpheta jamaicensis (leaf), and Carica papaya [24]. The above-stated compounds were extracted using hot water, cold water, and ethanol, which were reported to show good activity in the water-soluble region than ethanol. The aqueous and alcohol extracts of T. avicennioides, M. balsamina, C. paniculatum, and T. guineensis were effective against strains of multidrug-resistant S. typhi, whereas only the aqueous extracts of M. lucida and O. gratissimum were found to be active against MDR S. typhi [1]. Similarly, methanolic extract of Glycyrrhiza glabra and Azadirachta indica has been reported for anti-typhoid activity, broad-spectrum, anti-bacterial activity against gram-positive and gram-negative bacteria [21]. Likewise, eugenol (natural essential oil from clove) [11], resveratrol (flavonoid obtained from grapes, strawberries, cranberries, peanuts, and other plants) [13], and T. belerica extracts have been reported for bactericidal effect on S. typhi [14]. Besides, plant-derived compounds, bacterial by-products, or metabolites also have the potential anti-bacterial activity against S. typhi. The 3-phenyllactate (3-PLA), a metabolite produced from lactic acid bacteria (LAB) Lactobacillus plantarum ZJ316 in phenylalanine metabolism, is reported to exhibit anti-bacterial activity against Salmonella and also as an effective food preservative [23]. The 3-PLA is a novel anti-microbial organic acid with a broad spectrum against gram-positive, gram-negative bacteria and some fungi. They can impact pH, intracellular osmotic pressure, cell membrane permeability, material transport, cell signal transduction, energy metabolism, or biomacromolecule synthesis [28].
Previous studies have been reported that SSP extracted from marine algae shows potential anti-viral activity against human immunodeficiency virus (HIV), herpes simplex virus (HSV) and cytomegalovirus (CMV) [25]. Similarly, SSP extracted from Nothogeniu fastigiata also showed potent anti-viral activity against HIV, HCMV, HSV, and influenza virus [9]. The reports on marine-sulfated polysaccharide (MSP) extracted from the green macroalga Ulva armoricana have anti-bacterial activity against 42 bacterial strains including both gram-positive and gram-negative bacteria [3]. Further, anti-bacterial efficacy of fucoidan isolated from Sargassum wightii against the shrimp pathogen Vibrio parahaemolyticus and seven selected human bacterial pathogens were also reported [15], thus implying the capabilities of marine sulfated polysaccharides as a potential anti-microbial agent, particularly seaweed sulfated polysaccharides like carrageenans against Salmonella typhi infections. Thus, further in vitro and in vivo studies can be looked upon to take these bioactive SSPs to the next level of application in the crisis of XDR microbial diseases.
5 Conclusions
The frequent usage of classical antimicrobials has begun the increasing emergence of various drug-resistant pathogenic bacterial strains like MDR Salmonella typhi. In this context, the computational approaches for virtual screening of the natural sulfated polysaccharides of marine algae retrieved from the PubChem database were docked with the outer membrane protein F of Salmonella typhi to investigate the ability of the compounds to block the entry of the bacteria by preventing the attachment into the host. The docking results attained strongly suggest that the SSP had shown a better interaction with S. typhi OmpF equivalent to commercial leading drugs used for typhoid. Furthermore, among the SSP compounds, Carrageenan/MIV-150 showed the best binding affinity with H-bond interactions with drug-likeness properties. In addition, Carrageenan/MIV-150, β-carrageenan, and fucoidan compounds passed the ADMET criteria and highly progressed in all parameters. Hence, indicating the potentials of marine sulfated polysaccharides as a possible anti-microbial agent, particularly SSP-like carrageenans against Salmonella typhi infections. Therefore, based on the mechanism of interaction of these ligands with the OmpF target, it needs to be further evaluated by conducting in vitro studies to confirm their anti-microbial activity against the bacteria. These compounds would be the suitable drug candidates for the treatment of MDR S. typhi. Hence, these natural SSPs can be looked upon to the next level of application in the crisis of XDR microbial diseases.
Availability of data and materials
The data used and generated during the present study are obtainable from the corresponding author on reasonable request.
Abbreviations
- SSP:
-
Seaweed sulfated polysaccharide
- S. typhi :
-
Salmonella typhi
- OmpF:
-
Outer membrane porin F
- MDR:
-
Multiple drug resistance
- XDR:
-
Extensively drug resistance
- ADMET:
-
Absorption, distribution, metabolism, excretion, and toxicity
- CID:
-
Compound ID
- ADT:
-
AutoDockTools
- PDB:
-
Protein Data Bank
References
Akinyemi KO, Mendie UE, Smith ST, Oyefolu AO, Coker AO (2005) Screening of some medicinal plants used in south-west Nigerian traditional medicine for anti-Salmonella typhi activity. J Herb Pharmacother 5(1):45–60
Arunkumar M, Gunaseelan S, Kubendran Aravind M, Mohankumar V, Anupam P, Harikrishnan M et al (2021) Marine algal antagonists targeting 3CL protease and spike glycoprotein of SARS-CoV-2: a computational approach for anti-COVID-19 drug discovery. J Biomol Struct Dyn 25:1–28
Berri M, Slugocki C, Olivier M, Helloin E, Jacques I, Salmon H, Collen PN (2016) Marine-sulfated polysaccharides extract of Ulva armoricana green algae exhibits an anti-microbial activity and stimulates cytokine expression by intestinal epithelial cells. J Appl Phycol 28(5):2999–3008
Bredin J, Saint N, Malléa M, Dé E, Molle G, Pagès JM, Simonet V (2002) Alteration of pore properties of Escherichia coli OmpF induced by mutation of key residues in anti-loop 3 region. Biochem J 363(3):521–528
Bredin J, Simonet V, Iyer R, Delcour AH, Pagès JM (2003) Colicins, spermine and cephalosporins: a competitive interaction with the OmpF eyelet. Biochem J 376(1):245–252
Chatham-Stephens K, Medalla F, Hughes M, Appiah GD, Aubert RD, Caidi H, Nash J (2019) Emergence of extensively drug-resistant Salmonella typhi infections among travelers to or from Pakistan—United States, 2016–2018. Morb Mortal Wkly Rep 68(1):11
Cowan SW, Garavito RM, Jansonius JN, Jenkins JA, Karlsson R, König N, Schirmer T (1995) The structure of OmpF porin in a tetragonal crystal form. Structure 3(10):1041–1050
Crump JA, Luby SP, Mintz ED (2004) The global burden of typhoid fever. Bull World Health Organ 82:346–353
Damonte E, Neyts J, Pujol CA, Snoeck R, Andrei G, Ikeda S, Cerezo A (1994) Anti-viral activity of a sulfated polysaccharide from the red seaweed Nothogenia fastigiata. Biochem Pharmacol 47(12):2187–2192
Delcour AH (2009) Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta BBA Proteins Proteomics 1794(5):808–816
Devi KP, Nisha SA, Sakthivel R, Pandian SK (2010) Eugenol (an essential oil of clove) acts as an anti-bacterial agent against Salmonella typhi by disrupting the cellular membrane. J Ethnopharmacol 130(1):107–115
Gonzalez-Escobedo G, Marshall JM, Gunn JS (2011) Chronic and acute infection of the gall bladder by Salmonella typhi: understanding the carrier state. Nat Rev Microbiol 9(1):9–14
Lee W, Lee DG (2017) Resveratrol induces membrane and DNA disruption via pro-oxidant activity against Salmonella typhimurium. Biochem Biophys Res Commun 489(2):228–234
Madani A, Jain SK (2008) Anti-Salmonella activity of Terminalia belerica: in vitro and in vivo studies
Marudhupandi T, Kumar TTA (2013) Anti-bacterial effect of fucoidan from Sargassum wightii against the chosen human bacterial pathogens. Int Curr Pharm J 2(10):156–158
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67(4):593–656
Pages JM, James CE, Winterhalter M (2008) The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6(12):893–903
QikProp (2012) version 3.5, Schrödinger, LLC, New York, NY
Sanjeewa KA, Kang N, Ahn G, Jee Y, Kim YT, Jeon YJ (2018) Bioactive potentials of sulfated polysaccharides isolated from brown seaweed Sargassum spp in related to human health applications: a review. Food Hydrocoll 81:200–208
Simonet V, Malléa M, Pagès JM (2000) Substitutions in the eyelet region disrupt cefepime diffusion through the Escherichia coli OmpF channel. Antimicrob Agents Chemother 44(2):311–315
Somia G, Asma E, Zehra A (2015) An Alternative, H. T. Glycyrrhiza glabra and Azadirachta indica against Salmonella typhi: herbal treatment as an alternative therapy for typhoid fever
Steffen R, Rickenbach M, Wilhelm U, Helminger A, Schär M (1987) Health problems after travel to developing countries. J Infect Dis 156(1):84–91
Stoyanova LG, Ustyugova EA, Netrusov AI (2012) Anti-bacterial metabolites of lactic acid bacteria: their diversity and properties. Appl Biochem Microbiol 48(3):229–243
Varsha S, Agrawal RC, Sonam P (2013) Phytochemical screening and determination of anti-bacterial and anti-oxidant potential of Glycyrrhiza glabra root extracts. J Environ Res Dev 7(4A):1552
Witvrouw M, De Clercq E (1997) Sulfated polysaccharides extracted from sea algae as potential anti-viral drugs. Gener Pharmacol Vasc Syst 29(4):497–511
Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, Edwards DJ (2015) Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella typhi identifies inter-and intracontinental transmission events. Nat Genet 47(6):632–639
Wu SC (2017) Anti-oxidant activity of sulfated seaweeds polysaccharides by novel assisted extraction. In: Xu Z (ed) Solubility of polysaccharides. IntechOpen, London, pp 89–108
Zhou Q, Gu R, Li P, Lu Y, Chen L, Gu Q (2020) Anti-Salmonella mode of action of natural l-phenyl lactic acid purified from Lactobacillus plantarum ZJ316. Appl Microbiol Biotechnol 95:1–10
Ziervogel BK, Roux B (2013) The binding of antibiotics in OmpF porin. Structure 21(1):76–87
Acknowledgements
The authors thank the International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, for ADMET studies using Schrodinger software and DST-PURSE, DST-FIST, RUSA-MKU and UGC-UPE of Madurai Kamaraj University for facilities.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. The manuscript was written through contributions of all the authors, where PV designed the experiment; MA, MM, VAK, and MAK conducted the experiments; PV, VM, and BA analyzed the data; SG, MM, and PV wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Malaisamy Arunkumar, Vairamuthu Ashokkumar, and Sathaiah Gunaseelan are contributed equally to this work
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arunkumar, M., Mahalakshmi, M., Ashokkumar, V. et al. Evaluation of seaweed sulfated polysaccharides as natural antagonists targeting Salmonella typhi OmpF: molecular docking and pharmacokinetic profiling. Beni-Suef Univ J Basic Appl Sci 11, 8 (2022). https://doi.org/10.1186/s43088-021-00192-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43088-021-00192-x