Abstract
Background
HCV is a major risk factor for HCC; however, the exact mechanism of hepatocarcinogenesis is still not fully understood. Host genetic factors have been reported to play a significant role. Experimental studies support the tumor inhibitory effect of vitamin D on HCC cells. Several single nucleotide polymorphisms (SNPs) have been depicted in the vitamin D receptor (VDR) gene. We aimed to assess whether any of these polymorphisms could be significantly associated with increased risk of HCC.
Results
This study was conducted on 76 patients with HCV-related liver cirrhosis (48 patients had HCC on top of cirrhosis, and the other 28 had liver cirrhosis only). All patients underwent full medical history assessment, clinical examination, laboratory investigations, abdominal ultrasonography, and genotyping of the VDR gene. HCC patients had a significantly higher frequency of ApaI CC genotype compared with those patients without HCC. There is no statistically significant difference between the studied groups at any TaqI genotypes, but the carriage of the ApaI CC genotype had a significant association with liver disease severity in both patients groups compared with ApaI CA/AA genotypes. The carriage of the ApaI CC genotype was an independent predictor for HCC in HCV-related liver cirrhosis.
Conclusions
VDR ApaI polymorphism is significantly associated with the development of HCC; thus, ApaI CC genotype could be used as an important molecular marker to predict the risk of HCC in patients with HCV-related liver cirrhosis.
Similar content being viewed by others
Background
HCV is a major worldwide health problem. Globally, estimates indicate that about 71 million people are chronically infected [1]. The major complications of chronic HCV are hepatic fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [2].
HCC is considered to be the 6th common cancer all over the world, and it is accounting for 75 to 85% of primary liver cancers. HCC is in the fourth place as a cause of cancer death [3]. Besides HCV, there are a number of other risk factors that are suspected to cause HCC, including chronic HBV, non-viral cirrhosis, alcohol, non-alcoholic fatty liver disease (NAFLD), aflatoxin, hemochromatosis, Wilson’s disease, family history or genetic factors, and smoking [4].
The relationship between HCV and HCC is well known; however, the exact mechanism of carcinogenesis, including the host and viral factors, is not fully understood [5]. Different genetic factors are accused as a contributing factor for HCC in patients with chronic HCV, particularly gene polymorphisms of diverse inflammatory cytokines [6].
Vitamin D is a systemic hormone which is involved in the bone metabolism, but it also has a significant role in immunoregulation and cellular differentiation. Additionally, it has different anti-inflammatory and anticancer mechanisms through the vitamin D receptor (VDR) [7]. Studies have attributed the anticancer effect of vitamin D to its induction of cellular differentiation, proliferation, and angiogenesis inhibition, and hindering the progression of apoptosis as well [8].
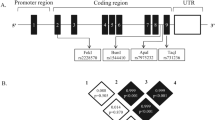
The VDR is a nuclear hormone receptor that combines with the active form of vitamin D (1,25(OH)2D3; calcitriol) and reacts with characteristic nucleotide sequences of target genes to perform its final biologic effects. The VDR gene is quite polymorphic and located on the 12q13.11 chromosome. Single nucleotide polymorphisms (SNPs) are frequently determined, and some of them are linked to carcinogenesis [9, 10].
Currently, data on VDR polymorphisms and their relationship with HCC are limited and extremely discordant [11]. As far as we know, there are a few published articles that have addressed such a relationship in Egyptian patients with HCV-related liver cirrhosis.
Methods
Seventy-six patients with HCV-related cirrhosis (56 males and 20 females, with age range 41–75 years) were included during the period from May 2017 to December 2018 in a cross-sectional study.
Forty-eight patients had HCC on top of liver cirrhosis [single or multiple more than 1 cm] (group HCC+) (36 males (75%), mean age 56.94 ± 6.82 years), while 28 patients had no HCC (group HCC−) (20 males (71.4%), mean age 57.5 ± 7.29 years). The study protocol was approved by the ethics committee of our institute. Informed written consent was obtained from all patients before inclusion in the study.
The exclusion criteria were the presence of other factors that could cause hepatocellular injury such as HBV co-infection (HBsAg-negative), history of alcoholism, autoimmune hepatitis (normal autoimmune markers; smooth muscle antibodies (SMA), anti-nuclear antibodies (ANA), and liver-kidney microsomal type 1 antibodies (LKM-1)), primary cholangitis (serum bilirubin and alkaline phosphatase levels, and MRCP in suspected cases), and primary biliary cirrhosis (negative anti-mitochondrial antibodies (AMA)).
All patients underwent complete medical history assessment, clinical examination, laboratory investigations, and abdominal ultrasonography. Abdominal triphasic CT scan was done if a hepatic focal lesion was detected on ultrasonography for the diagnosis of HCC.
The criteria for HCC diagnosis by CT were arterial phase enhancement pattern with rapid washout in the portal venous phase [12], liver disease severity estimated by Child-Turcotte-Pugh (CTP) classification [13], and model for end-stage liver disease (MELD) score [14].
Detection of VDR polymorphisms
PCR (polymerase chain reaction)
Genomic DNA was obtained from the buffy coat collected from EDTA blood using the QIAamp DNA Mini Kits (QIAGEN, Milan, Italy). The Ap fragment of the VDR gene containing the ApaI and TaqI restriction sites was amplified by PCR assay by using 200 ng of the genomic DNA in a total reaction volume of 50 μL. The PCR mix according to the manufacturer’s instructions consisted of 25 μL of master mix (Bioline, England) and 2.5 μL of each primer (10 pmol): forward (5′-CAGAGCATGGACAGGGAGCAA) and reverse (5′-GCAACTCCTCATGGCTGAGGTCTC)5. Thirty-five cycles of amplification were performed in a thermal cycler (T Gradient - Biometra). After the initial denaturation of DNA at 95 °C for 2 min, each cycle consisted of a denaturation step at 94 °C for 45 s, optimization of the primer annealing step modified to be at 58 °C for 1 min, an extension step at 72 °C for 1 min, and a final extension step at 72 °C for 7 min following the last cycle [15].
PCR products were analyzed on 2% agarose gel stained with ethidium bromide. The stained gels were visualized and analyzed with a gel documentation system to assess the size of PCR amplicon 745 bp.
RFLP assay (restriction fragment length polymorphism)
The amplified PCR products were then digested with the restriction endonucleases (ApaI and TaqI). For each endonuclease digestion reaction, 21.5 μL of the PCR product was digested with 1 μL (10 U) of the restriction enzyme ApaI (Jena Bioscience, Germany) or TaqI (Jena Bioscience, Germany), and 2.5 μL of restriction enzyme buffer “1×.” The resulting reaction solution (25 μL) was incubated at 37 °C for 1 h, then electrophoresed on 2% ethidium bromide-stained agarose gel, and visualized under UV illumination through a gel documentation system. Through a direct comparison with 100-bp DNA ladder (Jena Bioscience, Germany), the size of the DNA fragments was assessed. The restriction fragments generated after digesting the target gene by ApaI and TaqI restriction endonucleases are shown in Table 1.
Statistical analysis
Data were analyzed using the IBM SPSS Statistics for Windows, version 25 (IBM Corp., Armonk, NY, USA). Quantitative data were expressed as mean ± standard deviation, median, and range. Qualitative data were expressed as number and percentage. Quantitative data were tested for normality by the Shapiro–Wilk test. The Mann–Whitney U test and Kruskal–Wallis H test were used for data which were not normally distributed. Independent samples t test and one-way ANOVA test were used for normally distributed data. The chi-square (χ2) test and Fisher’s exact test were used for the comparison of qualitative variables as appropriate. Univariate and multivariate binary logistic regression analyses were used to determine the predictor variables of HCC. A 5% level was chosen as a level of significance in all statistical tests used in the study.
Results
The demographic variables, smoking status, comorbid illness, some laboratory data, Child class, and MELD score of the patient groups are summarized in Table 2.
The frequency distribution of VDR genotypes at ApaI and TaqI loci was summarized in Table 3. The HCC+ group had a statistically significant higher frequency of ApaI CC genotype compared to the HCC− group (P < 0.001). However, no statistically significant difference was found between the studied groups and any TaqI genotypes.
The digestion products of the VDR gene by ApaI and TaqI restriction enzymes were shown in Figs. 1, 2, 3, and 4.
Representative gel picture showing the PCR-RFLP analysis of ApaI VDR gene polymorphism on ethidium bromide-stained 2% agarose gel. M, marker (100 bp DNA ladder); lanes 4, 5, and 7 represent homozygous polymorphism (CC genotype; 531 and 214 bp bands); lanes 1, 2, 3, 6, 8, 9, and 13 represent no polymorphism (AA genotype; 745 bp band)
Representative gel picture showing the PCR-RFLP analysis of ApaI VDR gene polymorphism on ethidium bromide-stained 2% agarose gel. M, marker (100 bp DNA ladder); lanes 4, 6, and 7 represent heterozygous polymorphism (CA genotype; 745, 531, and 214 bp bands); lanes 1, 2, 3, and 5 represent no polymorphism (AA genotype; 745 bp band)
Representative gel picture showing the PCR-RFLP analysis of TaqI VDR gene polymorphism on ethidium bromide-stained 2% agarose gel. M, marker (100 bp DNA ladder); lanes 1, 2, and 3 represent heterozygous polymorphism (GA genotype; 495, 290, 245, and 205 bp bands); lane 4 represents no polymorphism (AA genotype; 495 and 245 bp bands)
The relation between the different ApaI genotypes and the severity of the liver disease among both groups is demonstrated in Table 4. In HCC− group, the carriage of the ApaI CC genotype was associated with a more severe liver disease (100% were Child C; MELD 20.33 ± 1.37; P = 0.011 and 0.01, respectively), compared to ApaI CA genotype (58.4% were Child C; MELD 17.92 ± 4.85) and ApaI AA genotype (10% were Child C; MELD 13.4 ± 4.69). Additionally, in the HCC+ group, the carriage of the ApaI CC genotype had a significant association with severe liver disease (52.6% were Child C vs. 0% for ApaI CA and AA genotypes; P = 0.003), ApaI CC carriers had the highest MELD score (P = 0.001). There is no significant difference between TaqI genotypes and liver disease severity among the studied groups.
Both univariate and multivariate binary logistic regression analyses confirmed that the carriage of the ApaI CC genotype (odds ratio (OR) 37.71, 95% confidence interval (CI95%) 5.83–244.12, P < 0.001) and platelet count (OR 1.02, CI95% 1.002–1.04, P = 0.01) were independent predictors for HCC development in patients with HCV-related liver cirrhosis (Table 5).
Discussion
The development of HCC is a complicated and multi-factorial process, in which both environmental and genetic factors play a role in carcinogenesis. The association between SNPs and HCC has been reported by numerous studies. These genetic traits may alter the natural history of cirrhosis and explain to some extent the observed differences in the risk of HCC development [16].
Interestingly, SNPs in the VDR gene are implicated in carcinogenesis in different organs such as the breast, prostate, skin, colon and rectum, and kidneys [17].
Here, we investigated the relationship between ApaI and TaqI VDR gene polymorphisms and HCC in patients with HCV-related liver cirrhosis.
We demonstrated that cirrhotic patients with HCC on top had a significantly higher frequency of VDR ApaI CC genotype compared to those patients who do not have HCC. Our results agree with previous studies [5, 15, 18, 19]. Some studies are matching us; they did not find a significant association between HCC and TaqI polymorphism [5, 15].
We investigated the implication of VDR ApaI and TaqI polymorphisms on the liver disease severity in both liver cirrhosis and HCC patients. The carriage of the ApaI CC genotype had significantly more severe liver disease (Child C and higher MELD score) compared to ApaI CA/AA genotypes, while the carriage of TaqI genotypes was not related to disease severity. These results came in agreement with Hung et al. [5] and Mohammed et al. [15].
On the contrary, Triantos et al. [20] reported that the carriage of VDR ApaI AA and TaqI AA genotypes was associated with more severe liver disease compared to ApaI CC/CA and TaqI GG/GA genotypes, respectively. This conflict could be simply explained by the difference in the inclusion criteria as Triantos et al. [20] investigated patients with chronic liver disease whatever the cause (viral, autoimmune, cryptogenic, etc.) and not complicated by HCC on top, while in our study, we selected only the HCV-related cirrhotic patients.
On addressing the risk factors for the development of HCC among HCV-related liver cirrhosis patients in this study, univariate binary logistic regression analysis was performed on age, sex, smoking, diabetes mellitus, Child and MELD scores, platelet count, and VDR ApaI and TaqI genotypes. Only platelet count and the carriage of the ApaI CC genotype were the factors significantly associated with HCC development; this was confirmed by multivariate binary logistic regression analysis.
The present study reported that the carriage of the ApaI CC genotype was an independent risk factor, proposing that the ApaI CC polymorphism may be a good molecular marker to predict the risk of HCC in patients with HCV-related liver cirrhosis. This agrees with Asal et al. [18] and El-Edel et al. [19]. Additionally, the ApaI CC genotype was reported to be associated with HCC development in non-cirrhotic patients with chronic HCV [5, 15].
Related studies of several polymorphisms in the VDR gene have been done to investigate their implication on the risk of HCC, although with different results. Falleti and his colleagues [21] demonstrated that the carriage of the BsmI GG and TaqI TT genotypes were significantly associated with HCC development in post-liver transplantation patients. However, this study was performed more particularly on patients with alcoholic cirrhosis, not other etiology of liver cirrhosis, where the carriage of the BAT [ATC] and [GTT] haplotypes was independently associated with an increased risk of HCC. This discrepancy could be explained by the low number of patients in the subgroup analysis of virus cirrhotic patients.
Peng et al. [10] reported that the carriage of the FokI TT/CT genotypes was associated with increased HBV-related HCC risk as compared to the FokI CC genotype. Some investigators in previous researches [15, 22,23,24] have reported that the carriage of the FokI TT genotype had a significantly higher risk for HCC after adjustment of other associated risk factors in those chronically infected with viral hepatitis. In addition, it was found that the FokI TT genotype was associated with advanced tumor stage and lymph node involvement.
On the contrary, Huang et al. [25] reported that VDR polymorphisms could influence the distinct clinical phenotypes in HBV carriers, but are not associated with HCC proposing a limited role of the VDR gene polymorphisms in carcinogenesis. However, a biochemical evidence obviously reported the inhibitory effect of vitamin D and its analogs on HCC cells [26]. Moreover, it had been described that the antiproliferative effect of vitamin D against malignant cells depends on the intracellular VDR level [27, 28].
Conclusions
In our country, the increasing HCC incidence is a result of the high prevalence of HCV, estimated to be around 14% in the general population [29, 30]. However, HCC is usually asymptotic, and diagnosis is usually made on an accidental basis. We suggested that the ApaI CC genotype may be used as a molecular marker to predict the risk of HCC in patients with HCV-related liver cirrhosis particularly in thrombocytopenic patients.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- HCV:
-
Hepatitis C virus
- HCC:
-
Hepatocellular carcinoma
- SNPs:
-
Several single nucleotide polymorphisms
- VDR:
-
Vitamin D receptor
- NAFLD:
-
Non-alcoholic fatty liver disease
- HBV:
-
Hepatitis B virus
- SMA:
-
Smooth muscle antibodies
- ANA:
-
Anti-nuclear antibodies
- LKM-1:
-
Liver-kidney microsomal type 1 antibodies
- MRCP:
-
Magnetic resonance cholangiopancreatography
- AMA:
-
Anti-mitochondrial antibodies
- CTP:
-
Child-Turcotte-Pugh
- MELD:
-
Model for end-stage liver disease
- PCR:
-
Polymerase chain reaction
References
The Polaris Observatory HCV Collaborators (2017) Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2(3):161–176
Hajarizadeh B, Grebely J, Dore GJ (2013) Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol 10(9):553–562
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB (2018) Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. AbdomRadiol (NY). 43(1):13–25
Hung CH, Chiu YC, Hu TH, Chen CH, Lu SN et al (2014) Significance of vitamin d receptor gene polymorphisms for risk of hepatocellular carcinoma in chronic hepatitis C. Transl Oncol 7(4):503–507
Bataller R, North KE, Brenner DA (2003) Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology 37(3):493–503
Raimondi S, Johansson H, Maisonneuve P, Gandini S (2009) Review and meta-analysis on vitamin D receptor polymorphisms and cancer risk. Carcinogenesis 30(7):1170–1180
Bikle D (2009) Nonclassic actions of vitamin D. J Clin Endocrinol Metab 94(1):26–34
Whitfield GK, Remus LS, Jurutka PW, Zitzer H, Oza AK, Dang HT et al (2001) Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol Cell Endocrinol 177(1-2):145–159
Peng Q, Yang S, Lao X, Li R, Chen Z, Wang J et al (2014) Association of single nucleotide polymorphisms in VDR and DBP genes with HBV-related hepatocellular carcinoma risk in a Chinese population. PLoS One 9(12):e116026
Louka ML, Fawzy AM, Naiem AM, Elseknedy MF, Abdelhalim AE, Abdelghany MA (2017) Vitamin D and K signaling pathways in hepatocellular carcinoma. Gene 629:108–116
Lencioni R, Cioni D, Della Pina C, Crocetti L, Bartolozzi C (2005) Imaging diagnosis. Semin Liver Dis. 25(2):162–170
Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R (1973) Transection of the oesophagus for bleeding oesophageal varices. Br J Surg 60(8):646–649
Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM, Kosberg CL et al (2001) A model to predict survival in patients with end-stage liver disease. Hepatology 33(2):464–470
Mohammed M, Omar N, Mohammed S, Deiab A (2017) The significance of vitamin D receptor gene polymorphisms for susceptibility to hepatocellular carcinoma in subjects infected with hepatitis C virus. Gastroenterol Hepatol Open Access 7(4):00246
Nahon P, Zucman-Rossi J (2012) Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol 57(3):663–674
Vaughan-Shaw PG, O’Sullivan F, Farrington SM, Theodoratou E, Campbell H, Dunlop MG et al (2017) The impact of vitamin D pathway genetic variation and circulating 25-hydroxyvitamin D on cancer outcome: systematic review and meta-analysis. Br J Cancer 116(8):1092–1110
Asal FES, El Bendary AS, El Khalawany WA, Abd-Elsalam S, Sheta BM (2017) The role of the vitamin D receptor gene polymorphisms in hepatocarcinogenesis in cirrhotic patients infected with chronic hepatitis C virus. Nat Sci 15(12):172–175
El-Edel RH, Mostafa MS, Montaser BA, Ali YAE-H (2017) Study of Apa-I vitamin D receptor gene polymorphism in patients with hepatocellular carcinoma. Menoufia Med J 30:619
Triantos C, Aggeletopoulou I, Kalafateli M, Spantidea PI, Vourli G, Diamantopoulou G et al (2018) Prognostic significance of vitamin D receptor (VDR) gene polymorphisms in liver cirrhosis. Sci Rep 8(1):14065
Falleti E, Bitetto D, Fabris C, Cussigh A, Fontanini E, Fornasiere E et al (2010) Vitamin D receptor gene polymorphisms and hepatocellular carcinoma in alcoholic cirrhosis. World J Gastroenterol. 16(24):3016–3024
Yao X, Zeng H, Zhang G, Zhou W, Yan Q, Dai L et al (2013) The associated ion between the VDR gene polymorphisms and susceptibility to hepatocellular carcinoma and the clinicopathological features in subjects infected with HBV. Biomed Res Int 2013:953974
Nada HA, Elsamanoudy AZ, Elalfy HA, Mogahed RE (2016) Study of vitamin D receptor-FOK-I gene polymorphism in chronic hepatitis C induced hepatocellular carcinoma patients: a case control study. Int J Innov Res Sci Eng Technol 5:4645–4655
Mohapatra S, Saxena A, Gandhi G, Koner BC, Ray PC (2013) Vitamin D and VDR gene polymorphism (FokI) in epithelial ovarian cancer in Indian population. J Ovarian Res. 6(1):37
Huang YW, Liao YT, Chen W, Chen CL, Hu JT, Liu CJ et al (2010) Vitamin D receptor gene polymorphisms and distinct clinical phenotypes of hepatitis B carriers in Taiwan. Genes Immun 11(1):87–93
Pourgholami M, Akhter J, Lu Y, Morris D (2000) In vitro and in vivo inhibition of liver cancer cells by 1, 25-dihydroxyvitamin D3. Cancer Lett 151(1):97–102
Dalhoff K, Dancey J, Astrup L, Skovsgaard T, Hamberg KJ, Lofts FJ et al (2003) A phase II study of the vitamin D analogue seocalcitol in patients with inoperable hepatocellular carcinoma. Br J Cancer 89(2):252–257
Merchan BB, Morcillo S, Martin-Nunez G, Tinahones FJ, Macías-González M (2017) The role of vitamin D and VDR in carcinogenesis: through epidemiology and basic sciences. J Steroid Biochem Mol Biol 167:203–218
El-Serag HB (2002) Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 35(5 Suppl 2):S72–S78
Hassany SM, Moustafa EFA, El Taher M, Abdeltawab AA, Blum HE (2015) Screening for hepatocellular carcinoma by Egyptian physicians. World J Gastrointest Oncol. 7(9):161–171
Acknowledgements
Not applicable
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
GMG, AA, and ANM collected, analyzed, and interpreted the data. NSA, NFF, and AS shared and helped in the designing and conceptualization of the study. EMA and UMA helped in the designing, editing, writing, and publishing of the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Sohag University Faculty of Medicine Ethical Committee (date 2018/2019; No. 1), and written informed consents were obtained from all participants. The procedures followed are in accordance with the institutional guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Galal, G.M., Abudeif, A., Ahmed, N.S. et al. Vitamin D receptor gene polymorphisms and risk of hepatocellular carcinoma in hepatitis C-related liver cirrhosis. Egypt Liver Journal 11, 3 (2021). https://doi.org/10.1186/s43066-020-00067-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43066-020-00067-3