Abstract
Background
Type 1 diabetes mellitus (T1D) results from environmental and genetic factors.
We aimed to investigate the distribution of PTPN22, IL2RA rs11594656, and rs2104286 variants and its association with T1D in children.
A case-control study conducted on 100 diabetic patients and 100 control children. PTPN22 gene, IL2RA rs11594656, and rs2104286 polymorphisms study were done by PCR followed by restriction fragment length polymorphism (RFLP) assay.
Results
T allele of PTPN22 gene was presented more frequently 47% in patient group versus 30% in controls, while C allele was 53% in the diabetic group versus 70% in controls showing a statistically significant difference between patient and control groups. Similarly, TT 1858 genotype was found in higher frequency with a statistically significant difference in favor of T1D patients (p = 0.038), OR (CI 95% 3.16 (1.28–7.09).
For IL2RA rs11594656 polymorphism, the frequency of TT, TA, and AA in patients at percentages of 20%, 60%, and 20% versus 4%, 60%, and 36% in controls respectively showed significant difference (p = 0.045). Also, T allele was detected more in patients group as evidenced by p = 0.059, OR (95% CI) of 2.38(1.49–6.12). Whereas, IL2RA rs2104286 polymorphism revealed a difference of otherwise non-statistical significance (p = 0.091). Those who harbored homozygous pattern of both IL2RA polymorphisms frequently had DKA and high mean HbA1C values.
Conclusion
PTPN22 (C1858T) and IL2RA rs11594656 polymorphisms increased the risk of T1DM development, while IL2RA rs2104286 polymorphism did not display any significant association among children with T1D. Having more than one risk allele could affect progression and control of T1D.
Similar content being viewed by others
Background
Type 1 diabetes mellitus (T1D) is a genetically complex disorder of glucose homeostasis that results from autoimmune destruction of the insulin-secreting cells of the pancreas. The development of T1D results from the interplay between environmental factors and several genes that participate in the susceptibility of this common metabolic disorder [1].
Genes for T1D may provide susceptibility to, or protection from, the disease. Although many chromosomal loci associated with such activities have been located, few true genes have been identified which can divide into 2 major subdivisions: human leukocyte antigen (HLA) genes and another major group named collectively as non-HLA genes. HLA is accounting for about 60% of genetic susceptibility for the disease. The HLA genes are a cluster of genes on chromosome 6 p 2 1[2]. The genes encode glycoproteins that are found on the surfaces of most cells and help the immune system to distinguish between self and non-self [3]. About 20 non-HLA loci contributing to disease susceptibility have been identified include the insulin gene, cytotoxic T-lymphocyte antigen-4 gene (CTLA-4), PTPN22 gene, and IL2RA gene [4].
(PTPN22) Protein tyrosine phosphatase non-receptor type 22 is a protein encoded by the PTPN22 gene located on the short arm of chromosome 1 (1p13.2) [4].
A C to T substitution at position 1858(C1858T) is a common mutation in the PTPN22 gene that results in the substitution of amino acid arginine with tryptophan at codon 620 (R620W), this single-nucleotide polymorphism (SNP) at position 1858 of PTPN22 was reported to be associated with T1D in many populations [5].
In human species, interleukin 2 receptor alpha (IL2RA) is a protein that is encoded by the IL2RA gene. This gene is located on the short arm of Chromosome10 (10p15.1).
However, IL2RA is a transmembrane protein of type 1 that is located on the surface of activated B cells, activated T cells, some myeloid precursors, oligodendrocytes, and thymocytes.
It has an essential role in the immune response of T cells and prevents autoimmune diseases with maintaining immune homeostasis [6].
It was suggested that single-nucleotide polymorphisms (SNPs) in the interleukin 2 receptor alpha (IL2RA) gene is associated with type T1D, including rs11594656, rs2104286, rs3118470, rs41295061, and rs706778.
In our study, the genetic risk factors for the development of T1D were the five SNPs in the IL2RA gene. However, rs11594656, rs2104286, and rs41295061 were the most associated SNPs [7].
Aim of the work
For these considerations of population variations, this study aimed to investigate the relation of PTPN22 1858 C/T gene polymorphism and T1D in a random group of Egyptian children known to be diabetic. Also, the association between IL2RA rs11594656 and rs2104286 polymorphisms and T1D within the same group.
Methods
The present study was carried out on 100 Egyptian children with T1D (group I) from our pediatric department and endocrinology clinic at the Menoufia University Hospitals, Egypt, including 38 males and 62 females; their ages ranged from 2 years to 16 years with variable duration of illness and degree of diabetes control, diagnosed according to the criteria of American Diabetes Association (ADA).
The exclusion criteria
Children with T2D with secondary diabetes or those with primary renal illness were excluded from the study.
Equal number of age- and sex-matched Egyptian children, who had negative family history of auto-immune disorders and normal glucose parameters, were taken as control group (group II). A local ethical committee for a research study approved the study (approval number: 53631373). The ethical guidelines of the 1964 Declaration of Helsinki and its later amendments were followed during the study.
After taking informed consent, all patients and controls were subjected to the following:
Detailed history taking including family pedigree, complete examination of all body systems including anthropometric measurements was done followed by molecular study of PTPN22 gene polymorphism and that of IL2RA rs11594656, rs2104286 polymorphisms were analyzed by PCR-RFLP, and the procedure was completed through these steps:
DNA extraction using DNA was extracted by DNA extraction kit (Gene JET Whole Blood Genomic DNA Purification Mini Kit) according to the manufacturer’s instructions. As a second station, polymerase chain reaction (PCR) where genomic DNA was amplified using PCR after adjustment of thermal cycler conditions for which different sets of specific primers were used as follows; forward and reverse primers for amplification of PTPN22 C1858T polymorphism were; forward primer: 5′-ACTGATAATGTTGCTTCAACGG-3′ and reverse primer: 5′-TCACCAGCTTCCTCAACCAC-3′ to be ready for digestion with RsaI. The enzyme-digested product was analyzed by 1.5% agarose gel electrophoresis after ethedium bromide staining to be visualized under UV. Upon recognition of its specific targeted site of the amplified sequence, the enzyme cuts the C allele into 2 fragments of 172 bp and 46 bp, while the T allele remain intact (218 bp).
Genotyping of IL2RA SNPs rs11594656 polymorphism was done by using the following primers; forward primer: 5-CTCCCCAGTCATTCACCAAA-3 and the reverse primer: 5-TCTTTTGGCTTTTCTCACTATGATGCCGTC-3, and then, Bsm AI enzyme-digest product was electrophoresed on a 1.5% agarose gel. The digested A allele was cut into 2 fragments of 211 bp and 101 bp, while the T allele remain intact 312 bp. For genotyping of IL2RA SNPs rs2104286 polymorphism, forward primer: 5-GCAGGTGTCAACGCAAAAAC-3 and Reverse primer TCCCTGGAATGTCACTGATG -3 were applied followed by visualization of NdeI-digested product on 1.5% agarose gel, yielding a digested G allele cut into 2 fragments, while the A allele remained intact (288 bp).
All the above steps of molecular analysis were done also in the genetic lab of our Genetic and Endocrinology Unit, Pediatric Department, Faculty of Medicine, Menoufia University Hospitals.
Statistical analysis
All data were tabulated then statistically analyzed through SPSS 20.0 (SPSS Inc., Chicago, IL, USA), expressed as mean ± SD. The difference between the studied parameters in the two groups was assessed via Student’s t test. The frequencies were actually expressed in percentages (%), the differences in genotyping and allele frequencies in between the studied groups were assessed by chi-square (χ2). Probability (P) was considered of statistical significance at values less than 0.05 [8].
Results
The study included 100 patients with T1D (group I), 47 males (47%) and 53 females (53%) and their ages ranged from 2 to 16 years with a mean age of 8.48 ± 4.07. Equal number of healthy children, matched for age and sex were taken as controls. Forty-five (45%) patients had a family history of diabetes mellitus, 55(55%) patients were without a family history of diabetes. Of the diseased children, 82(82%) showed negative consanguinity. Baseline characteristics and laboratory data of studied groups at the time of the study were shown in (Table 1). According to their clinical aspects, 64% of diabetic children presented with DKA. However, 36% presented with classic symptoms. Regarding FBS, 2h/PP, and HbA1c, there were statistically significant difference between patient and control groups (P <0.05), while the mean of cholesterol, LDL, triglyceride, and thyroid function (TSH, FT4) did not show any significant changes between T1D patient and control subjects (P >0.05).
In respect to PTPN22 gene, the genotypic results revealed that 5(5%) had TT, 84(84%) TC, and 11(11%) had CC for the patient group, while in the control group the genotypic results were 0%, 60%, and 40% for TT, TC, and CC variants respectively, and these results showed a statistically significant difference between patient and control groups (p = 0.028).
As for allelic distribution, frequency of T allele was more (47%) in the diabetic group versus (30%) in controls, while C allele was 53% in the diabetic group versus 70% in controls, with a statistically significant difference (P = 0.038), OR [CI 95% 3.16 (1.28–7.09)].
The frequency distribution of IL2RA rs11594656 for TT, TA, and AA differed significantly in patients 20%, 60%, and 20% versus 4%, 60%, and 36% in controls respectively (p = 0.045). Moreover, T allele was presented frequently in T1D patients than in control individuals; p = 0.059, OR (CI 95%) 2.38(1.49–6.12).
On the other hand, IL2RA rs2104286 showed higher frequency number of A allele (75%) in the group of patients with T1D versus (78%) in controls in comparison to G allele, but this difference was not of statistical significance (p = 0.089) (Table 2).
While considering the relation between demographic and clinical data of patients to the genotypic variants of PTPN22, it showed no significant difference regarding sex distribution. Similar results were observed in relation to age at onset of the disease, duration of T1D and pattern of presentation (Table 3).
Of much interest, grouping of patients revealed that there were 11 of them had homozygous pattern of both IL2RA rs11594656 and IL2RA rs2104286 polymorphisms with the following characteristics: their ages at onset of diabetes ranged from 3 to 11 years with mean of 6.65 ± 3.93. Duration of illness was 4.5–10 years with a mean of 3.75 ± 1.99 years. Positive consanguinity was reported in only 2 (22.2%) patients. However, 8 (72.6%) patients had a family history of diabetes mellitus. Furthermore, those patients presented recurrent attacks of DKA in 9 (81.8%) patients. HbA1C was 8.80–12.30 with a mean of 10.77 ± 1.73; a reliable indicator of poor control of T1D (Table 4).
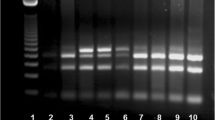
Distribution of electrophoretic bands of PTPN22 C1858T, IL2RA rs11594656, and IL2RA rs2104286 genotype in patients with T1D and control subjects was shown in Figs. 1, 2, and 3 in order.
Distribution of in PTPN22 C1858T genotype in diabetic patients and controls. Genotyping of PTPN22 from left: • Lane 1: DNA ladder 100bp. Band (T) at (218) bp Band (C) two fragments at (172) bp and (46) bp • Lane 2, and 8: Homozygous (CC genotype) only (C) band present. • Lane 3, 4, 5, 6, and 7: Heterozygous (TC) where both T band and C band
Distribution of in IL2RA rs11594656 genotypes in diabetic patients and controls. Genotyping of IL2RA rs11594656 genotypes from left: • Lane 1: DNA ladder 100bp. Band (T) at (312) bp Band (A) two fragments at (211) bp and (101) bp • All Lanes showing: Homozygous (AA genotype) only (A) band present as two fragments at 211bp and 101 bp
Distribution of in IL2RA rs2104286 genotypes in diabetic patients and control samples. Genotyping of IL2RA rs2104286 genotypes from left: • Lane 1: DNA ladder 100bp. Band (A) at (288) bp Band (G) appears as two fragments. • Lanes 2, 3, 4, 5, and 7: Heterozygous (AG) where both A band and G band • Lane 6,and 8 :Homozygous (GG genotype)
Discussion
Susceptibility to T1D is determined by complex interactions between several genetic loci and environmental factors. Many of them harbor functional candidates likely involved in autoimmunity and potentially in T1D pathogenesis, and relevant examples are PTPN22 and IL2RA genes [9,10,11].
In our study, we found a statistically significant difference between patient and control groups regarding family history (p < 0.001). This finding was in agreement with Pociot et al. [12] who reported that the risk to develop T1D increases to1 in 20 in a population who have the first-degree relative with T1DM, while the risk among the general population is 1 in 300.
In our study, the distribution of PTPN22 (C1858T) polymorphisms revealed that the distribution of the homozygous pattern (TT) was a 5% in patient group versus 0% in the control group, while heterozygous pattern (TC) was 84% inpatient group versus 60% in the control group. On the other hand, wild variant (CC) was 11% in patient group versus 40% in the control group, all these results of genotypic variation showed a statistically significant difference between patients and controls (p = 0.028). These findings were obvious while analyzing the allelic distribution of high frequency of T allele that was of statistical significance between the studied groups.
Our genotypic results were in agreement with Bottini et al. [13], who firstly reported an association between the PTPN22 polymorphism and T1D from North America and Sardinia. On the frame of the necessity of throughout additional research to clarify the varied geographic distribution, recent several studies involved different populations had explored the relationship between PTPN22 and T1M including those from Italy [14, 15], Spain [16], Denmark [17], Finland [18], Brazil [19], France [20], Germany [21, 22], the UK [23], Poland [24], Greece [25], China [26], and Egypt [27]. On the contrary, other studies were reported which denied the association between 1858C/T polymorphism and susceptibility to T1D [28, 29].
These contradictory results can be explained by the limited size of the sample, genetic heterogeneity among the studied populations, the different environmental factors involved in the pathogenesis of T1D, or other methodological issues.
Regarding the association between gender, age of onset of T1D, type of presentation, and PTPN22 C1858T in T1D patients, in our study, there was statistically insignificant difference between PTPN22 genotypes (TT, TC, and CC) and the above mentioned parameters (P > 0.05); these results agreed with some studies [26, 28]. On the other hand, they were in disagreement with others who found gender differentiation in favor of females [27, 30].
In the present study, the distribution of IL2RA rs11594656 gene polymorphisms revealed that TT, TA, and AA in T1DM patients were frequently observed at ratio of 20%, 60%, and 20% respectively versus 4%, 60%, and 36% in controls with a statistically significant difference (p = 0.045). Obviously, a predominance of TT genotype was seen in patients than controls, and these findings agreed with the studies which reported that IL2RA rs11594656 (TT genotype) as a risk of developing T1D [7, 24, 27, 31], compared to others with opposing results [32,33,34].
Although our results had considered (TT) genotype of IL2RA rs11594656 a significant risk factor of developing T1D regardless of the patients' clinical presentation, reviewing data from past literature showed association of T allele with younger age of onset of T1D [24, 30,31,32,33,34,35,36,37].
While analyzing the distribution of IL2RA rs2104286 gene polymorphisms, findings of the current study showed genotype frequencies as follows: AA, AG, and GG in T1DM patients was 60%, 36%, and 4% respectively versus 57%, 36%, and 7% in controls; these results were of non-statistical significance as shown by p = 0.091. The same was evident as regards the low frequency of A allele of IL2RArs2104286 polymorphism p = 0.86, OR [CI 95% 0.48 (0.19–1.18)]. Our results were found to be in agreement with some related research studies who reported that the studied polymorphism did not display any significant association with T1D [24, 32] in contrast to others that provided evidence for the association between T1D and rs2104286 [7, 31, 37]. Population differences and diversity in the influence of genetic/environmental factors could be claimed for this discrepancy.
In the current study, 11 patients representing 11% of the whole sample of T1D patients were detected to have a homozygous pattern of both IL2RA rs11594656 and IL2RA rs2104286 polymorphisms characterized by positive family history of diabetes (72.6%), poor diabetes control with HbA1C values ranged 8.80–12.30% and mean values of 10.77 ± 1.73, in addition to documented history of recurrent attacks of DKA in 81.8%. These findings may indicate that the conjunction of the homozygous pattern of both IL2RA rs11594656 and IL2RA rs2104286 polymorphisms played an important role in the degree of glycemic control of T1D with the need for further efforts paved towards better life expectancy of those children based on the fact that realized the polygenic nature of T1DM [38, 39].
Although previous studies reported the association between PTPN22 1858 C/T SNP and many autoimmune diseases, Elsisi et al. denied any further evidence that the PTPN22 gene may play a role in the genetic susceptibility to T1D in Egyptian children [28].
However, a recent Egyptian study reported that T allele of PTPN22 gene and TT genotype of IL2RA were associated with T1D [27].
This finding was prominent in female patients and those with early onset diabetes. Our study is considered accumulated evidence for the use of these analyzed genes as biomarkers for T1D susceptibility among Egyptian children.
The study was conducted in a relatively small number of patients; so further studies on a larger sample of patients are recommended to confirm our finding. Also, the effect of PTPN22 (C1858T) and IL2RA rs11594656 polymorphisms on diabetes-related antibodies, control of the disease, and insulin requirements should be elucidated in further studies.
Conclusions
Our study suggests that PTPN22 and IL2RA rs11594656 polymorphism increase the risk of T1D, while IL2RA rs2104286 polymorphism showed a non-significant difference for association with T1D. Our findings can be a baseline step for the clustering strategies approximated for the use of these analyzed genes as genetic biomarkers for T1D susceptibility among Egyptian children and adolescents.
Availability of data and materials
None to be declared.
Abbreviations
- ADA:
-
American Diabetes Association
- CI:
-
Confidence interval
- CTLA-4:
-
Cytotoxic T-lymphocyte antigen-4 gene
- DKA:
-
Diabetic ketoacidosis
- FBS:
-
Fasting blood sugar
- FT4:
-
Free thyroxin
- HbA1c:
-
Hemoglobin A1C
- HLA:
-
Human leukocyte antigen
- IL2RA:
-
Interleukin-2 receptor alpha
- LDL:
-
Low-density lipoprotein
- OR:
-
Odds ratio
- PCR:
-
Polymerase chain reaction
- PTPN22:
-
Protein tyrosine phosphatase non-receptor type 22
- SNPs:
-
Single-nucleotide polymorphisms
- T1D:
-
Type 1 diabetes mellitus
- TSH:
-
Thyroid-stimulating hormone
References
American Diabetes Association (2019) Classification and diagnosis of diabetes: standards of medical care in diabetes—2019. Diabetes Care 42(1):S13–S28
Ilonen J, Kiviniemi M, Lempainen J, Simell O, Toppari J, Veijola R, Knip M, Finnish Pediatric Diabetes Register (2016) Genetic susceptibility to type 1 diabetes in childhood–estimation of HLA class II-associated disease risk and class II effect in various phases of islet autoimmunity. Pediatr Diabetes 17:8–16
Nigrovic PA, Raychaudhuri S, Thompson SD (2018) Genetics and the classification of arthritis in adults and children. Arthritis Rheum 70(1):7–17
Wallace DJ (2019) The lupus book: a guide for patients and their families. Oxford University Press
Stanford SM, Bottini N (2014) PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol 10(10):602
Huang X, Kühne V, Kun JF, Soboslay PT, Lell B, Velavan TP (2012) In-vitro characterization of novel and functional regulatory SNPs in the promoter region of IL2 and IL2R alpha in a Gabonese population. BMC Med Genet 13(1):117
Tang W, Cui D, Jiang L, Zhao L, Qian W, Long SA, Xu K (2015) Association of common polymorphisms in the IL 2 RA gene with type 1 diabetes: evidence of 32,646 individuals from 10 independent studies. J Cell Mol Med 19(10):2481–2488
Levesque R (2007) SPSS programming and data management: a guide for SPSS and SAS users, 4th edn. SPSS Inc., Chicago
Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nierras CR, Todd JA, Rich SS, Nerup J (2010) Genetics of type 1 diabetes: what's next? Diabetes 59(7):1561–1571
Smyth DJ, Cooper JD, Howson JM, Walker NM et al (2008) PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes. 57:1730–1737
Stene LC, Rønningen KS, Bjørnvold M, Undlien DE et al (2010) An inverse association between history of childhood eczema and subsequent risk of type 1 diabetes that is not likely to be explained by HLA-DQ, PTPN22, or CTLA4 polymorphisms. Pediatr Diabetes 11:386–393
Pociot F, Lernmark Å (2016) Genetic risk factors for type 1 diabetes. Lancet 387(10035):2331–2339
Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T (2004) A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36:337–338
Blasetti A, Di Giulio C, Tumini S, Provenzano M, Rapino D, Comegna L et al (2017) Role of the C1858T polymorphism of protein tyrosine phosphatase non-receptor type 22 (PTPN22) in children and adolescents with type 1 diabetes. Pharm J 17:186–191
Saccucci P, Del DE, Rapini N, Verrotti A et al (2008) Association between PTPN22 C1858T and type 1 diabetes: a replication in continental Italy. Tissue Antigens 71:234–237
Santiago JL, Martínez A, de la Calle H, Fernández-Arquero M et al (2007) Susceptibility to type 1 diabetes conferred by the PTPN22 C1858T polymorphism in the Spanish population. BMC Med Genet 8:54
Nielsen C, Hansen D, Husby S, Lillevang ST (2007) Sex-specific association of the human PTPN22 1858T-allele with type 1 diabetes. Int J Immunogenet 34:469–473
Hermann R, Lipponen K, Kiviniemi M, Kakko T et al (2006) Lymphoid tyrosine phosphatase (LYP/PTPN22) Arg620Trp variant regulates insulin autoimmunity and progression to type 1 diabetes. Diabetologia 49:1198–1208
Chagastelles PC, Romitti M, Trein MR, Bandinelli E et al (2010) Association between the 1858T allele of the protein tyrosine phosphatase nonreceptor type 22 and type 1 diabetes in a Brazilian population. Tissue Antigens 76:144–148
Chelala C, Duchatelet S, Joffret ML, Bergholdt R et al (2007) PTPN22 R620W functional variant in type 1 diabetes and autoimmunity related traits. Diabetes 56:522–526
Dultz G, Matheis N, Dittmar M, Röhrig B et al (2009) The protein tyrosine phosphatase non-receptor type 22 C1858T polymorphism is a joint susceptibility locus for immunthyroiditis and autoimmune diabetes. Thyroid 19:143–148
Kahles H, Ramos-Lopez E, Lange B, Zwermann O et al (2005) Sex-specific association of PTPN22 1858T with type 1 diabetes but not with Hashimoto’s thyroiditis or Addison’s disease in the German population. Eur J Endocrinol 153:895–899
Smyth D, Cooper JD, Collins JE, Heward JM et al (2004) Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 53:3020–3023
Fichna M, Zurawek M, Fichna P, Januszkiewicz D, Nowak J (2012) Polymorphic variants of the IL2RA gene and susceptibility to type 1 diabetes in the polish population. Tissue Antigens 79:198–203
Giza S, Goulas A, Gbandi E, Effraimidou S, Papadopoulou-Alataki E, Eboriadou M, Galli-Tsinopoulou A (2013) The role of PTPN22 C1858T gene polymorphism in diabetes mellitus type 1: first evaluation in greek children and adolescents. Biomed Res Int. 2013; 2013:721604. https://doi.org/10.1155/2013/721604. Epub 2013 Jul 15.
Liu HW, Xu RY, Sun RP, Wang Q, Liu JL, Ge W, Yu Z (2015) Association of PTPN22 gene polymorphism with type 1 diabetes mellitus in Chinese children and adolescents. Genet Mol Res 14(1):63–68
Abdelrahman HM, Sherief LM, Elrahman DM, Alghobashy A, Elsaadani HF, Mohamed RH (2016) The association of PTPN22 (rs2476601) and IL2RA (rs11594656) polymorphisms with T1D in Egyptian children. Hum Immunol 77(8):682–686
Elsisi O, Kamal M, Madani H, Ibrahim A, Elsheikh S (2015) Association of protein tyrosine phosphatase non-receptor type 22 (PTPN22) C1858T gene polymorphism with type 1 diabetes mellitus in Egyptian children cohort. Egypt Pediat Assoc Gazette 63(3-4):75–79
Baniasadi V, Das SN (2008) No evidence for association of PTPN22 R620W functional variant C1858T with type 1 diabetes in Asian Indians. J Cell Mol Med 12:1061–1062
Kordonouri O, Hartmann R, Badenhoop K, Kahles H, Ilonen J (2010) PTPN22 1858T allele is associated with younger age at onset of type 1 diabetes and is not related to subsequent thyroid autoimmunity. Hum Immunol 71:731–732
Belot MP, Fradin D, Mai N, Le Fur S, Zelenika D, Kerr-Conte J, Pattou F, Lucas B, Bougneres P (2013) CpG methylation changes within the IL2RA promoter in type 1 diabetes of childhood-onset. PLoS One. 8(7):68093. https://doi.org/10.1371/journal.pone.0068093.
Qu HQ, Bradfield JP, Bélisle A, Grant SFA, Hakonarson H, Polychronakos C (2009) The type I diabetes association of the IL2RA locus. Genes Immun 10(1):S42–S48
Maier LM, Lowe CE, Cooper J, Downes K, Anderson D, Severson A (2009) IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet 5:e1000322
Kawasaki TA, Ikegami H, Kobayashi T, Maruyama T, Nakanishi K et al (2009) IL2RA and type 1 diabetes in Japanese. J Clin Endocrinol Metab 94(3):947–952
Klinker MW, Schiller JJ, Magnuson VL, Wang T, Basken J, Veth K, Pearce KI, Kinnunen L, Harjutsalo V, Wang X, Tuomilehto J (2010) Single-nucleotide polymorphisms in the IL2RA gene are associated with age at diagnosis in late-onset Finnish type 1 diabetes subjects. Immunogenetics 62(2):101–107
Aminkeng F, Weets I, Van Autreve JE, Koeleman BP, Quartier E, Van Schravendijk C, Gorus FK, Van der Auwera BJ, Belgian Diabetes Registry (2010) Association of IL-2RA/CD25 with type 1 diabetes in the Belgian population. Hum Immunol 71(12):1233–1237
Howson JM, Walker NM, Smyth DJ, Todd JA (2009) Analysis of 19 genes for association with type I diabetes in the type I diabetes genetics consortium families. Genes Immun 10(1):S74–S84
Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH (2013) Projecting the performance of risk prediction based on polygenic analysis studies. Nat Genet 45(4):400–405 e1-3
Udler MS, McCarthy MI, Florez JC, Mahajan A (2019) Genetic risk score for diabetes diagnosis and precision medicine. Endocr Rev 40(6):1500–1520
American Diabetes Association (2018a) Diagnosis and classification of diabetes mellitus. Diabetes Care 31(1):S55–S60
Acknowledgements
No acknowledgements.
Funding
None.
Author information
Authors and Affiliations
Contributions
NFB conceived the study. SSA, MAT, and ZSA participated in its design and coordination. NFB and MAT provided key technical guidance. SSA, ZSA, and NFB analyzed and interpreted the patient data. NFB, MAT drafted the manuscript, and SSA and critically revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures performed in the study were in accordance with the ethical standards of our institutional research committee, Faculty of Medicine, Menoufia University. The approval number was 58637385.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abou El Ella, S.S., Tawfik, M.A., Mohammed, Z.S. et al. PTPN22 gene and IL2RA rs11594656, rs2104286 gene variants: additional insights of polygenic single-nucleotide polymorphisms’ pattern among Egyptian children with type 1 diabetes. Egypt Pediatric Association Gaz 69, 35 (2021). https://doi.org/10.1186/s43054-021-00079-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43054-021-00079-7