Abstract
Background
Insulin-like growth factor 2 (IGF2) mRNA-binding proteins 2 (IGF2BP2/IMP2), an RNA-binding protein encoded by the IGF2BP2 gene, exerts its influence across diverse pathological pathways. While accumulating evidence underscores the potential significance of IGF2BP2 in the tumorigenesis of specific cancers, a comprehensive pan-cancer investigation into its role remains absent.
Methods
Consequently, we conducted an exhaustive exploration employing a multitude of databases to elucidate the plausible oncogenic implications of IGF2BP2. This encompassed a comprehensive scrutiny of its expression profiles, prognostic implications, association with cancer-associated fibroblast infiltration, biological functionality in distinct tumor contexts, and plausible correlations with drug sensitivities.
Results
Our findings showed that IGF2BP2 was highly expressed in some types of cancers, but presented at low levels in several cancer types. Furthermore, the role of IGF2BP2 in predicting prognosis exhibited a dichotomous interplay across varied cancer types. Remarkably, observations unveiled the cancer-associated fibroblast infiltration within specific tumors, notably encompassing breast invasive carcinoma of the luminal A subtype, kidney renal clear cell carcinoma, ovarian serous cystadenocarcinoma, pheochromocytoma and paraganglioma, and prostate adenocarcinoma, and thymoma. Intriguingly, gene enrichment analyses spotlighted the co-expression of IGF2BP2 with genes implicated in pivotal biological processes, including DNA replication and recombinational repair.
Conclusion
Our investigation intricately unveils the potential of IGF2BP2 as a versatile prognostic biomarker across diverse tumor categories. This study bridges existing knowledge gaps and augments the understanding of IGF2BP2’s intricate involvement in tumorigenesis, underscoring its significance as a prospective avenue for therapeutic intervention.
Similar content being viewed by others
Introduction
Cancer stands as the foremost cause of mortality worldwide [1], posing a formidable challenge to both life quality and longevity. Regrettably, a comprehensive cure for cancer remains elusive. To broaden the knowledge of potential therapeutic strategies for malignancies, a better understanding of carcinogenic mechanisms and cancer progression for the targeting oncogenes is crucial. The advent of extensive multi-omics datasets, exemplified by the Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) [2, 3], empowers the execution of pan-cancer expression analyses. These analyses uncover commonalities, distinctions in gene expression profiles, and their associations with clinical outcomes [4].
Amidst this context, the human insulin-like growth factor 2 (IGF2) mRNA-binding proteins 2 (IGF2BP2/IMP2) encoded by the IGF2BP2 gene which is located on chromosome 3q27, emerges as a multifaceted RNA-binding protein orchestrating a plethora of biological processes [5]. IGF2BP2, with a molecular mass of 66 kDa, is an oncofetal protein important for embryonic development and downregulated in normal adult tissues, which functions by binding and stabilizing mRNA to extend their half-life [6]. The multifunctional role of IGF2BP2 has been substantiated by a series of studies highlighting its participation in vital cellular functions, encompassing cell polarization, migration, morphology, metabolism, proliferation, and differentiation [7]. Recent insights reveal IGF2BP2's role as an N6-methyladenosine (m6A) reader, interacting with diverse RNA species to influence the genesis and progression of cancers [8]. Beyond this, IGF2BP2 emerges as an autonomous prognostic determinant across a spectrum of cancer types, encompassing acute myelocytic leukemia [9], breast cancer [10], colorectal cancer [11], hepatocellular carcinoma [12], oral squamous cell carcinoma [13], pancreatic cancer [14], thyroid cancer [15], and many others. Moreover, IGF2BP2's influence extends to metabolic disorders [16], such as diabetes [17] and obesity [18].
Nevertheless, prevailing investigations concerning the role of IGF2BP2 have predominantly centered on individual cancer types. To our present knowledge, a comprehensive inquiry into the functional and clinical significance of IGF2BP2 across the spectrum of pan-cancer remains conspicuously absent. In this pursuit, we methodically harness multiple databases to undertake an exhaustive analysis encompassing IGF2BP2's expression profiles, prognostic implications, cancer-associated fibroblast infiltration, biological functions in the context of pan-cancer, and its plausible correlations with drug sensitivities. In synthesis, our findings unequivocally present IGF2BP2 as a prospective prognostic indicator across an array of malignancies. Notably, IGF2BP2 exerts its function via tumor microenvironment (TME) and exhibits discernible associations with drug sensitivities. This comprehensive study unlocks novel perspectives on IGF2BP2's overarching role in pan-cancer scenarios, offering potential therapeutic avenues for exploration.
Materials and methods
Gene expression analysis of IGF2BP2
Initially, an IGF2BP2 mRNA expression plot was meticulously plotted utilizing the Human Protein Atlas (HPA) database (version 21) (https://www.proteinatlas.org/) [19]. To elucidate gene conservation across vertebrates, a comprehensive visualization was facilitated through the UCSC genome browser (http://www.genome.ucsc.edu/cgi-bin/hgTracks) [20]. In tandem, UCSC Xena datahubs (http://xena.ucsc.edu/) were used to obtain RNA-seq datasets from the TCGA [21] and the GTEx [3] repositories for a pan-cancer differential expression of IGF2BP2. After data processing, there were 10,534 samples from TCGA and 7568 samples from GTEx included for analysis. Furthermore, within the purview of the Tumor Immune Estimation Resource version 2 (TIMER2) framework (http://timer.cistrome.org/) [22], the “Gene_DE” module was employed to scrutinize variations in IGF2BP2 expression between tumor and adjacent normal tissues across diverse malignancies.
Correlation of IGF2BP2 expression with survival prognosis
The investigation encompassed the correlation between IGF2BP2 mRNA expression and clinical endpoints, namely overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI). To discern these relationships, we employed Cox proportional hazard regression models and Kaplan–Meier survival analyses. For these analyses, the R software (version 4.2.2) integrated the “survival,” “survminer,” and “ggplot2” packages. A two-sided P value less than 0.05 was considered statistically significant.
Cancer-associated immune infiltration analysis
Using the “Immune” module of TIMER2.0 [23], Extended Polydimensional Immunome Characterization (EPIC), Microenvironment Cell Populations-counter (MCPCOUNTER), XCELL, and the Tumor Immune Dysfunction and Exclusion (TIDE) algorithms were employed to investigate the correlation between IGF2BP2 expression and cancer-associated fibroblast infiltration. Spearman’s rank correlation test was used to calculate partial correlation coefficients (cor) and associated P values. The discerned outcomes were portrayed via a heatmap and scatter plots.
IGF2BP2-related gene enrichment analysis
The “Similar Gene Detection” module within GEPIA2 [24] was used to extract the top 100 IGF2BP2-associated target genes from the TCGA tumor datasets. These genes exhibited patterns closely resembling IGF2BP2 expression. Subsequently, the gene ontology (GO) pathway enrichment analysis was performed in the R package “clusterProfiler” grounded on the aforementioned 100 genes. The results encompassing biological process (BP), cellular component (CC), and molecular function (MF) were portrayed through a bubble chart.
The STRING tool (version 11.5) (https://string-db.org/) was used to establish a homo sapiens IGF2BP2 co-expression network [25]. This endeavor adhered to these parameters: (1) network type: full STRING network; (2) meaning of network edges: evidence; (3) active interaction sources: co‑expression; (4) minimum required interaction score: low confidence (0.150); and (5) max number of interactors to show: no more than 50 interactors.
Drug sensitivity analysis of IGF2BP2
For an intricate insight into the drug sensitivity of IGF2BP2 across pan-cancer contexts, we engaged two curated datasets labeled “RNA: RNA-seq” and “Compound activity: DTP NCI-60.” These datasets, procured from the CellMiner™ (Version 2022.3) [26], were pivotal in scrutinizing the responsiveness of IGF2BP2 to diverse compounds (https://discover.nci.nih.gov/cellminer/home.do). To ensure the analytical robustness, drugs approved by FDA or clinical trials were selected for analysis, which was processed with the R-packages “impute,” “limma,” “ggplot2,” and “ggpubr.” The most significant 16 outputs were presented in scatter plots.
Results
Expression of IGF2BP2 in pan-cancer
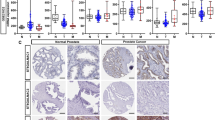
Within the confines of HPA and GTEx datasets, IGF2BP2 emerged as a gene characterized by modest tissue specificity, while demonstrating enrichment in retinal, small intestinal, and placental contexts (Fig. 1A). Furthermore, it was found that IGF2BP2 was relatively conserved among vertebrates (Fig. 1B). Then, the expression of IGF2BP2 in pan-cancer was further explored through the RNA-seq data obtained from TCGA and GTEx databases. Except for those cancers without available normal tissue data, significant expression differences of IGF2BP2 were noted in 28 types of cancers. Notably, IGF2BP2 exhibited pronounced overexpression in cancers such as cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), and uterine carcinosarcoma (UCS) (Fig. 1C). Conversely, reduced IGF2BP2 levels were discerned in tumor tissues relative to normal counterparts in cancers including adrenocortical carcinoma (ACC), breast invasive carcinoma (BRCA), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), kidney renal clear cell carcinoma (KIRC), brain lower-grade glioma (LGG), lung adenocarcinoma (LUAD), pheochromocytoma and paraganglioma (PCPG), and prostate adenocarcinoma (PRAD) (Fig. 1C). For the augment of our perspective, we sought the expression blueprint of IGF2BP2 in human pan-cancer scenarios through the TIMER database. This exploration illuminated the upregulation of IGF2BP2 in CHOL, COAD, ESCA, GBM, HNSC, KIRP, LIHC, LUSC, STAD, and THCA. Conversely, IGF2BP2 was downregulated in BRCA, KIRC, PCPG, and PRAD (Fig. 1D). These findings collectively posited IGF2BP2 as a potential promoter of carcinogenesis in some types of cancers, prompting further clinical exploration.
Differential expression of IGF2BP2. A Consensus IGF2BP2 tissue expression based on datasets of HPA (Human Protein Atlas) and GTEx (Genotype-Tissue Expression). B IGF2BP2 gene conservation analysis among vertebrates visualized using the UCSC genome browser. C Differential IGF2BP2 mRNA expression between TCGA tumors and GTEx normal tissues. D IGF2BP2 mRNA expression in different tumor types in TIMER2. *p < 0.05, **p < 0.01, and ***p < 0.001
Pan-cancer prognostic implications for IGF2BP2
To unveil the intricate interplay between IGF2BP2 expression levels and prognosis, rigorous Cox proportional hazards modeling and Kaplan–Meier analysis were executed across diverse cancers. Our findings underscored that elevated IGF2BP2 expression was significantly linked to shortened OS in the cases of BLCA (p = 0.005), HNSC (p = 0.018), LAML (p = 0.010), KIRC (p = 0.023), LGG (p < 0.0001), LUAD (p = 0.013), MESO (p = 0.013), and PAAD (p = 0.006), while tied to prolonged OS in UVM (p = 0.003) (Fig. 2). Furthermore, disease-specific survival (DSS) analysis divulged that escalated IGF2BP2 expression correlated with unfavorable outcomes in cases of BLCA (p = 0.009), KIRC (p < 0.0001), LGG (p < 0.0001), LUAD (p = 0.046), MESO (p = 0.012), and PAAD (p = 0.004), while better outcome for patients with UVM (p = 0.001) (Fig. 3). Additionally, progression-free interval (PFI) analysis uncovered that augmented IGF2BP2 expression signaled a less favorable prognosis for BLCA (p = 0.034), KIRC (p < 0.001), LGG (p < 0.0001), LUAD (p = 0.023), and PCPG (p = 0.029), while aligning with a better prognosis for THYM (p = 0.017) (Fig. 4). In aggregate, these observations linked heightened IGF2BP2 expression with compromised prognosis across diverse cancer categories.
Correlation between IGF2BP2 expression and immune infiltration
Recent investigations have highlighted the pivotal role of cancer-associated fibroblasts (CAFs) within the intricate tumor microenvironment (TME), influencing the regulation of tumor-infiltrating immune cells (TIICs) that significantly shape cancer progression [27, 28]. In light of this, we employed algorithms including EPIC, MCPCOUNTER, XCELL, and TIDE to unravel potential connections between CAF infiltration levels and IGF2BP2 expression across varying cancer types. Strikingly, our findings consistently indicated statistically significant positive associations between IGF2BP2 expression and CAF infiltration in BRCA-LumA, KIRC, OV, PCPG, PRAD, and THYM, as delineated through these four algorithmic approaches (Fig. 5).
IGF2BP2-related gene enrichment analysis
To unravel the functional mechanisms underpinning the role of IGF2BP2 in cancer genesis and progression, we extracted and integrated the top 100 genes displaying expression profiles akin to IGF2BP2 across TCGA tumor datasets. The consequential gene ontology (GO) enrichment analysis yielded a host of genes linked to cellular biology of DNA including DNA replication, DNA-dependent DNA replication, recombinational repair, and more (Fig. 6A). These results were then cross-validated by the STRING tool, identifying a spectrum of 50 genes co-expressed with IGF2BP2, thus corroborating the findings of the GO analysis. The interplay within this network of genes is depicted in Fig. 6B.
Drug sensitivity of IGF2BP2 in pan-cancer
The potential correlation between drug sensitivity and IGF2BP2 was further explored using the CellMiner™ database. Notably, IGF2BP2 expression was negatively correlated with drug sensitivity of dexrazoxane, etoposide, SR16157, teniposide, raloxifene, M-AMSA, fulvestrant, XK-469, ribavirin, zalcitabine, bendamustine, idarubicin, nitrogen mustard, tamoxifen, idoxuridine, and mitoxantrone (Fig. 7). This observation underscored the potential linkage between IGF2BP2 and chemoresistance to various therapeutic agents, including widely used treatments such as teniposide and ribavirin, commonly employed in clinical contexts.
Drug sensitivity analysis of IGF2BP2. The expression of IGF2BP2 was associated with the sensitivity of Dexrazoxane (A), Etoposide (B), SR16157 (C), Teniposide (D), Raloxifene (E), M-AMSA (F), Fulvestrant (G), XK-469 (H), Ribavirin (I), Zalcitabine (J), Bendamustine (K), Idarubicin (L), Nitrogen mustard (M), Tamoxifen (N), IDOXURIDINE (O), and Mitoxantrone (P)
Discussion
The fusion of bioinformatics advancements and comprehensive molecular investigations has facilitated the discernment of molecular biomarkers and their intricate roles in pan-cancer scenarios [29, 30]. In this study, we embarked on an analytical journey to illuminate the multifaceted role of IGF2BP2 in the context of oncogenesis and prognostication across diverse malignancies.
IGF2BP2, co-existing within the IGF2BP family alongside IGF2BP1 and IGF2BP3, constitutes a highly conserved cluster of RNA-binding oncofetal proteins pivotal in RNA stability, localization, and translation. Originally identified in 1999, IGF2BP2 anchors itself to the 5′ untranslated regions (5′UTRs) of the translationally modulated IGF-II reader mRNA [31]. Its structure involves two RNA-recognizing motifs (RRMs) at the N-terminal and four human heterogeneous nuclear ribonucleoprotein (hnRNP)-K homology (KH) domains at the C-terminal [32]. While early investigations predominantly focus on IGF2BP2's correlation with type 2 diabetes susceptibility [17], recent years have seen a surge in studies spotlighting its aberrant expression's nexus with tumor onset and progression. Notably, studies involving IGF2BP2 knockout mice substantiated its pivotal role in malignant tumor advancement [33]. Additionally, IGF2BP2's engagement in colorectal cancer proliferation and survival via modulating RAF-1 degradation through miR-195 was highlighted by Ye et al. [34], while Mu et al. [35] pointed out its promotion of GBM progression via PI3K/AKT pathway activation through IGF2 regulation. Although these insights were invaluable, a comprehensive assessment of IGF2BP2's implications across diverse tumor types remained uncharted territory. As such, we embarked on a systematic characterization of IGF2BP2 across varied malignancies, evaluating its distinctive characteristics encompassing gene expression, prognostic implications, and immune infiltration.
Our findings corroborated previous studies, showcasing IGF2BP2's pervasive expression across diverse tissues with modest specificity, yet indicating pronounced presence within a spectrum of tumors. Aligned with prior research, similar results were observed in cases of COAD, ESCA, GBM, HNSC, LAML, LIHC, and PAAD [9, 11, 12, 14, 36,37,38,39]. However, discrepancies emerged, such as the contrasting discovery of IGF2BP2's overexpression in BRCA accompanied by an augmented autoimmune response [40]. Plausible explanations for these disparities could encompass variations in study demographics, breast cancer subtypes, and genetic polymorphism [10, 41, 42]. Notably, divergent trends in IGF2BP2 expression between LUSC and LUAD warranted attention. The insights into IGF2BP2's restraint of NSCLC cell proliferation and invasion underscored the nuanced roles within distinct NSCLC classifications [43, 44]. This nuanced interplay within lung cancer subtypes signaled the necessity for granular subtype-based analyses.
Our investigation delved deeper into the connection between elevated IGF2BP2 expression and prognostic outcomes. Survival analysis underscored that high IGF2BP2 expression aligned with unfavorable prognosis in BLCA, HNSC, KIRC, LAML, LGG, LUAD, MESO, PAAD, and PCPG. This corroborated findings offered by He et al. [9] who conducted a meta-analysis, establishing IGF2BP2 overexpression as a predictor of poorer OS in LAML patients [HR = 1.31(1.16–1.49); p = 0.00]. In parallel, Xu et al. found IGF2BP2 was an independent predictor of adverse prognosis in pancreatic cancer [HR = 2.395 (1.655–4.134); p < 0.05] [14]. Notably, Barghash et al. [41] identified IGF2BP2 overexpression as a hallmark of basal-like breast cancer associated with shorter survival, unveiling its diverse implications across cancer subtypes. Intriguingly, our study revealed an unconventional scenario, wherein high IGF2BP2 expression correlated with favorable prognosis in UVM and THYM. The uniqueness of these rare cancers precluded reference from existing studies. The paradox of the opposing effects of IGF2BP2 expression may be due to the context-specific manner by which IGF2BP2 regulates cellular processes through different regulatory networks. Elevated levels of IGF2BP2 may promote antitumor immunity by mediating the infiltration of immunocytes, thereby contributing to favorable prognosis [45].
An evolving body of evidence underscored the potential of TME characteristics as predictive biomarkers for immunotherapy responsiveness and clinical outcomes [46]. Our observations suggested a positive correlation between IGF2BP2 expression and infiltration of CAFs in select tumor types. CAFs, pivotal stromal components, secrete growth factors, inflammatory mediators, and extracellular matrix (ECM) proteins critically implicated in tumor initiation, progression, and metastasis [47]. Recent work had demonstrated that CAFs were associated with worse prognosis, therapeutic resistance, and disease recurrence [48, 49]. It was also reported that IGF2BP2 had an effect on various immune cell subtypes, hinting at its potential role in hepatocellular carcinoma (HCC) therapy [50]. Collectively, these results underscored IGF2BP2's intricate role in cancer immunity and its broad prognostic implications.
Our exploration uncovered a network of genes co-expressed with IGF2BP2 across diverse tumors and tissues, facilitated through GEPIA2. Furthermore, gene enrichment analyses underscored IGF2BP2's potential impact on cancer etiology, particularly in DNA replication and recombinational repair processes. These data were consistent with previously published articles, indicating that the genes regulated by IGF2BP2 were mainly enriched in cell proliferation [9]. Intriguingly, recent research implicated the role of IGF2BP3 in DNA replication during the cell cycle [51], alluding to potential functional interplay considering their co-expression. Such insights paved the way for in-depth investigations into the molecular functions of IGF2BP2.
Past decades witnessed intensive research on IGF2BP1 and 3 as promising targets for chemotherapy development, with small-molecule inhibitors emerging [7, 52]. Our findings aligned IGF2BP2 with chemoresistance to multiple agents, which was a critical challenge in cancer therapy. As reported, IGF2BP2 induced chemoresistance in GBM cells by inhibiting FOXO1-mediated PID1 expression and promoted glioma progression [53]. A recent breakthrough highlighted the potential of a small-molecule compound (CWI1-2) for IGF2BP2 inhibition, displaying promising anti-leukemia effects [54]. This convergence of evidence underscored IGF2BP2's clinical significance and its potential as a therapeutic target, warranting rigorous exploration for future anti-cancer drug design.
While our study contributed new dimensions to the understanding of IGF2BP2's impact on diverse tumors, certain limitations deserved attention. Sample size constraints for uncommon cancers might introduce bias or batch effects. Our work provided preliminary insights into IGF2BP2's role in various cancers, yet further in vitro and in vivo experimentation is imperative to elucidate its precise biological function and underlying carcinogenic mechanisms. As the journey into IGF2BP2's complexities continues, our findings lay the groundwork for informed, targeted therapeutic strategies for the benefit of cancer patients.
Conclusion
To summarize, our investigation underscores the prevalent overexpression of IGF2BP2 across a spectrum of cancers, rendering it a promising candidate for prognostic biomarkers in select cancer types. This study unveils the intricate and varied involvement of IGF2BP2 in the realm of pan-cancer, thereby furnishing a compelling basis for refining therapeutic strategies to precisely target IGF2BP2, aligning with the goal of personalized treatment approaches.
Availability of data and materials
The data of the current study are available from the following open public databases: TCGA (https://cancergenome.nih.gov) and GTEx (https://gtexportal.org/home/datasets) as is described above. Other data will be obtained from the corresponding authors upon reasonable request.
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- BP:
-
Biological process
- BRCA:
-
Breast invasive carcinoma
- CAFs:
-
Cancer-associated fibroblasts
- CC:
-
Cellular component
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL:
-
Cholangiocarcinoma
- COAD:
-
Colon adenocarcinoma
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- DSS:
-
Disease-specific survival
- ECM:
-
Extracellular matrix
- EPIC:
-
Extended polydimensional immunome characterization
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- GO:
-
Gene ontology
- GTEx:
-
The Genotype-Tissue Expression
- HCC:
-
Hepatocellular carcinoma
- hnRNP:
-
Heterogeneous nuclear ribonucleoprotein
- HNSC:
-
Head and neck squamous cell carcinoma
- HPA:
-
Human Protein Atlas
- IGF2:
-
Insulin-like growth factor 2
- IGF2BP2/IMP2:
-
Insulin-like growth factor 2 mRNA-binding proteins 2
- KH:
-
K homology
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Brain lower-grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MCPCOUNTER:
-
Microenvironment cell populations-counter
- MF:
-
Molecular function
- OS:
-
Overall survival
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PFI:
-
Progression-free interval
- PRAD:
-
Prostate adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- RRMs:
-
RNA-recognizing motifs
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach adenocarcinoma
- TCGA:
-
The Cancer Genome Atlas
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TIDE:
-
Tumor Immune Dysfunction and Exclusion
- TIICs:
-
Tumor-infiltrating immune cells
- TIMER2:
-
Tumor Immune Estimation Resource version 2
- TME:
-
Tumor microenvironment
- UCS:
-
Uterine carcinosarcoma
- UTRs:
-
Untranslated regions
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Blum A, Wang P, Zenklusen JC (2018) SnapShot: TCGA-analyzed tumors. Cell 173(2):530. https://doi.org/10.1016/j.cell.2018.03.059
Consortium GT (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45(6):580–585. https://doi.org/10.1038/ng.2653
Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD (2020) Pan-cancer analysis of whole genomes. Nature 578(7793):82–93. https://doi.org/10.1038/s41586-020-1969-6
Cao J, Mu Q, Huang H (2018) The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int 2018:4217259. https://doi.org/10.1155/2018/4217259
McMullen ER, Gonzalez ME, Skala SL, Tran M, Thomas D, Djomehri SI et al (2018) CCN6 regulates IGF2BP2 and HMGA2 signaling in metaplastic carcinomas of the breast. Breast Cancer Res Treat 172(3):577–586. https://doi.org/10.1007/s10549-018-4960-2
Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M et al (2013) Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci: CMLS 70(15):2657–2675. https://doi.org/10.1007/s00018-012-1186-z
Wang J, Chen L, Qiang P (2021) The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int 21(1):99. https://doi.org/10.1186/s12935-021-01799-x
He X, Li W, Liang X, Zhu X, Zhang L, Huang Y et al (2018) IGF2BP2 overexpression indicates poor survival in patients with acute myelocytic leukemia. Cel Physiol Biochem: Int J Exp Cell Physiol Biochem Pharmacol 51(4):1945–1956. https://doi.org/10.1159/000495719
Liu G, Zhu T, Cui Y, Liu J, Liu J, Zhao Q et al (2015) Correlation between IGF2BP2 gene polymorphism and the risk of breast cancer in Chinese Han women. Biomed Pharmacother = Biomed Pharmacothe 69:297–300. https://doi.org/10.1016/j.biopha.2014.12.017
Cui J, Tian J, Wang W, He T, Li X, Gu C et al (2021) IGF2BP2 promotes the progression of colorectal cancer through a YAP-dependent mechanism. Cancer Sci 112(10):4087–4099. https://doi.org/10.1111/cas.15083
Pu J, Wang J, Qin Z, Wang A, Zhang Y, Wu X et al (2020) IGF2BP2 promotes liver cancer growth through an m6A-FEN1-dependent mechanism. Front Oncol 10:578816. https://doi.org/10.3389/fonc.2020.578816
Wang X, Xu H, Zhou Z, Guo S, Chen R (2022) IGF2BP2 maybe a novel prognostic biomarker in oral squamous cell carcinoma. Biosci Rep. https://doi.org/10.1042/bsr20212119
Xu X, Yu Y, Zong K, Lv P, Gu Y (2019) Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J Exp Clin Cancer Res: CR 38(1):497. https://doi.org/10.1186/s13046-019-1470-y
Dong L, Geng Z, Liu Z, Tao M, Pan M, Lu X (2021) IGF2BP2 knockdown suppresses thyroid cancer progression by reducing the expression of long non-coding RNA HAGLR. Pathol Res Pract 225:153550. https://doi.org/10.1016/j.prp.2021.153550
Dai N (2020) The diverse functions of IMP2/IGF2BP2 in metabolism. Trends Endocrinol Metab 31(9):670–679. https://doi.org/10.1016/j.tem.2020.05.007
Christiansen J, Kolte AM, Hansen T, Nielsen FC (2009) IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol 43(5):187–195. https://doi.org/10.1677/jme-09-0016
Wu HH, Liu NJ, Yang Z, Tao XM, Du YP, Wang XC et al (2014) IGF2BP2 and obesity interaction analysis for type 2 diabetes mellitus in Chinese Han population. Eur J Med Res 19(1):40. https://doi.org/10.1186/2047-783x-19-40
Ponten F, Schwenk JM, Asplund A, Edqvist PH (2011) The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med 270(5):428–446. https://doi.org/10.1111/j.1365-2796.2011.02427.x
Lee BT, Barber GP, Benet-Pages A, Casper J, Clawson H, Diekhans M et al (2022) The UCSC genome browser database: 2022 update. Nucleic Acids Res 50(D1):D1115–D1122. https://doi.org/10.1093/nar/gkab959
Zhang Z, Li H, Jiang S, Li R, Li W, Chen H et al (2019) A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data. Brief Bioinform 20(4):1524–1541. https://doi.org/10.1093/bib/bby023
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS et al (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Can Res 77(21):e108–e110. https://doi.org/10.1158/0008-5472.CAN-17-0307
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q et al (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48(W1):W509–W514. https://doi.org/10.1093/nar/gkaa407
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47(W1):W556–W560. https://doi.org/10.1093/nar/gkz430
von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31(1):258–261. https://doi.org/10.1093/nar/gkg034
Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J et al (2012) Cell Miner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Can Res 72(14):3499–3511. https://doi.org/10.1158/0008-5472.can-12-1370
Chen X, Song E (2019) Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 18(2):99–115. https://doi.org/10.1038/s41573-018-0004-1
Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL (2019) Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol 16(5):282–295. https://doi.org/10.1038/s41575-019-0115-0
Chen F, Fan Y, Cao P, Liu B, Hou J, Zhang B et al (2021) Pan-cancer analysis of the prognostic and immunological role of HSF1: a potential target for survival and immunotherapy. Oxid Med Cell Longev 2021:5551036. https://doi.org/10.1155/2021/5551036
Xu WX, Zhang J, Hua YT, Yang SJ, Wang DD, Tang JH (2020) An integrative pan-cancer analysis revealing LCN2 as an oncogenic immune protein in tumor microenvironment. Front Oncol 10:605097. https://doi.org/10.3389/fonc.2020.605097
Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC (1999) A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19(2):1262–1270. https://doi.org/10.1128/mcb.19.2.1262
Liu HB, Muhammad T, Guo Y, Li MJ, Sha QQ, Zhang CX et al (2019) RNA-binding protein IGF2BP2/IMP2 is a critical maternal activator in early zygotic genome activation. Adv Sci 6(15):1900295. https://doi.org/10.1002/advs.201900295
Dai N, Zhao L, Wrighting D, Krämer D, Majithia A, Wang Y et al (2015) IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab 21(4):609–621. https://doi.org/10.1016/j.cmet.2015.03.006
Ye S, Song W, Xu X, Zhao X, Yang L (2016) IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Lett 590(11):1641–1650. https://doi.org/10.1002/1873-3468.12205
Mu Q, Wang L, Yu F, Gao H, Lei T, Li P et al (2015) Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol Ther 16(4):623–633. https://doi.org/10.1080/15384047.2015.1019185
Lu F, Chen W, Jiang T, Cheng C, Wang B, Lu Z et al (2022) Expression profile, clinical significance and biological functions of IGF2BP2 in esophageal squamous cell carcinoma. Exp Ther Med. https://doi.org/10.3892/etm.2022.11177
Janiszewska M, Suvà ML, Riggi N, Houtkooper RH, Auwerx J, Clément-Schatlo V et al (2012) Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev 26(17):1926–1944. https://doi.org/10.1101/gad.188292.112
Barghash A, Golob-Schwarzl N, Helms V, Haybaeck J, Kessler SM (2016) Elevated expression of the IGF2 mRNA binding protein 2 (IGF2BP2/IMP2) is linked to short survival and metastasis in esophageal adenocarcinoma. Oncotarget 7(31):49743–49750. https://doi.org/10.18632/oncotarget.10439
Deng X, Jiang Q, Liu Z, Chen W (2020) Clinical significance of an m6A reader gene, IGF2BP2, in head and neck squamous cell carcinoma. Front Mol Biosci. https://doi.org/10.3389/fmolb.2020.00068
Liu W, Li Y, Wang B, Dai L, Qian W, Zhang JY (2015) Autoimmune response to IGF2 mRNA-binding protein 2 (IMP2/p62) in breast cancer. Scand J Immunol 81(6):502–507. https://doi.org/10.1111/sji.12285
Barghash A, Helms V, Kessler SM (2015) Overexpression of IGF2 mRNA-binding protein 2 (IMP2/p62) as a feature of basal-like breast cancer correlates with short survival. Scand J Immunol 82(2):142–143. https://doi.org/10.1111/sji.12307
Almawi WY, Zidi S, Sghaier I, El-Ghali RM, Daldoul A, Midlenko A (2022) Novel association of IGF2BP2 gene variants with altered risk of breast cancer and as potential molecular biomarker of triple negative breast cancer. Clin Breast Cancer. https://doi.org/10.1016/j.clbc.2022.12.017
Huang RS, Zheng YL, Li C, Ding C, Xu C, Zhao J (2018) MicroRNA-485-5p suppresses growth and metastasis in non-small cell lung cancer cells by targeting IGF2BP2. Life Sci 199:104–111. https://doi.org/10.1016/j.lfs.2018.03.005
Han L, Lei G, Chen Z, Zhang Y, Huang C, Chen W (2021) IGF2BP2 regulates MALAT1 by serving as an N6-methyladenosine reader to promote NSCLC proliferation. Front Mol Biosci 8:780089. https://doi.org/10.3389/fmolb.2021.780089
Cao Y, Jiao N, Sun T, Ma Y, Zhang X, Chen H et al (2021) CXCL11 correlates with antitumor immunity and an improved prognosis in colon cancer. Front Cell Dev Biol 9:646252. https://doi.org/10.3389/fcell.2021.646252
Wu T, Dai Y (2017) Tumor microenvironment and therapeutic response. Cancer Lett 387:61–68. https://doi.org/10.1016/j.canlet.2016.01.043
Biffi G, Tuveson DA (2021) Diversity and biology of cancer-associated fibroblasts. Physiol Rev 101(1):147–176. https://doi.org/10.1152/physrev.00048.2019
Chen Y, McAndrews KM, Kalluri R (2021) Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 18(12):792–804. https://doi.org/10.1038/s41571-021-00546-5
Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM et al (2020) A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 20(3):174–186. https://doi.org/10.1038/s41568-019-0238-1
Wei Q (2021) Bioinformatical identification of key genes regulated by IGF2BP2- mediated RNA N6-methyladenosine and prediction of prognosis in hepatocellular carcinoma. J Gastrointest Oncol 12(4):1773–1785. https://doi.org/10.21037/jgo-21-306
Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B (2020) RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res: CR 39(1):203. https://doi.org/10.1186/s13046-020-01714-8
Wallis N, Oberman F, Shurrush K, Germain N, Greenwald G, Gershon T et al (2022) Small molecule inhibitor of Igf2bp1 represses Kras and a pro-oncogenic phenotype in cancer cells. RNA Biol 19(1):26–43. https://doi.org/10.1080/15476286.2021.2010983
Han J, Yu X, Wang S, Wang Y, Liu Q, Xu H et al (2021) IGF2BP2 induces U251 glioblastoma cell chemoresistance by inhibiting FOXO1-mediated PID1 expression through stabilizing lncRNA DANCR. Front Cell Dev Biol 9:659228. https://doi.org/10.3389/fcell.2021.659228
Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y et al (2022) The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell 40(12):1566–82.e10. https://doi.org/10.1016/j.ccell.2022.10.004
Acknowledgements
We would like to thank Dr. Chi Zhang for English language editing.
Funding
This work was supported by the Science and Technology Innovation Plan of Shanghai Science and Technology Commission [Grant No. 21S11909900].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by H-LZ and D-DC. The first draft of the manuscript was written by H-LZ, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
As these databases are open-source and informed consent of patients is not required, ethics committee approval by the institutional review board was not needed for this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, HL., Chen, DD. & Li, XL. Pan-cancer analysis identified IGF2BP2 as a potential prognostic biomarker for multiple tumor types. Egypt J Med Hum Genet 25, 3 (2024). https://doi.org/10.1186/s43042-023-00468-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-023-00468-0