Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent responsible for the onset of coronavirus disease 2019 (COVID-19), elicits a wide range of clinical manifestations, spanning from asymptomatic infection to the development of severe and potentially fatal diseases. The involvement of host microribonucleic acids (microRNAs) in the cytokine storm induced by SARS-CoV-2 infection has been observed, leading to their identification as potential biomarkers for COVID-19. Consequently, the objective of this study was to examine the expression of miRNA-20a and miRNA-320 in 145 COVID-19 patients and 145 healthy controls using quantitative real-time polymerase chain reaction (qRT-PCR).
Results
The results indicated a significant increase in miRNA-20a expression with fold change (10.65; P ≤ 0.0001), while decreased expression of miRNA-320 was found with fold change (0.48; P ≤ 0.0001) in COVID-19 patients. The specificity and sensitivity of miRNAs in COVID-19 patients were assessed using receiver operating characteristic (ROC) analysis.
Conclusion
miRNA-20a and miRNA-320 may serve as potential COVID-19 diagnostic biomarkers with high sensitivity and specificity.
Similar content being viewed by others
Background
The single-stranded RNA pathogens known as coronaviruses (CoVs) are responsible for gastrointestinal, pulmonary, hepatic, and central nervous system disorders in both humans and animals [1]. Human cells may be infected by six distinct types of CoV, such as SARS-CoV, HCoVOC43, HKU-NL63, HCoV-HKU1, HCoV-229E, and MERS-CoV [2]. All coronaviruses have a similar genomic structure, with an open reading frame (ORF) 1a/b at the 5′ end that codes for 16 non-structural proteins (NSP1-NSP16) and four structural proteins that code by ORF at the 3′ end (the envelope (E), nucleocapsid (N), membrane (M) and spike (S) proteins). For the SARS-CoV-2 and SARS-CoV viruses to infect host cells, transmembrane serine protease 2 (TMPRSS2), the S protein must interact with the proteins of the extracellular membrane of the host [3, 4].
The novel coronavirus disease 2019 (COVID-19), caused by the new coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has speedily spread throughout the world after the first case was reported in late December 2019 in Wuhan, China. As of December 1, 2020, there will be 1,528,984 COVID-19-related mortality and 66,243,918 confirmed sick patients, according to the (WHO) [5]. A great deal of clinical and academic interest has been generated by the recognition of the biochemical and molecular mechanisms underlying SARS-CoV-2 infection and disease formation in human host cells in light of the global health crisis. Finding biomarkers that could be used as the disease's diagnostic and treatment targets is a crucial goal.
One of the indicators of the miRNA family may change the targets of genes related to immunity through networks of virus-host cell interactions [6]. Small noncoding RNA molecules, or miRNAs, are naturally occurring molecules that attach to specific mRNA targets through nucleotide similarity. By inhibiting translation or causing the target mRNAs to degrade, these chemicals contribute to the regulation of gene expression in a variety of organisms [7]. Viral infections and pathogenic processes, as well as chronic disorders, are all correlated with dysregulated miRNA expression [8, 9].
Cellular miRNAs can directly hinder viral replication within host cells, although the precise mechanisms underlying their interaction with viruses are still not entirely known. Several techniques have been proposed, including stopping viral replication and blocking viral genome translation, which can result in an anti-viral response by deregulating the synthesis of numerous endogenous miRNAs. miRNAs' anti-viral response may include modulating their mRNA targets: WNT, INF, MAPK, PIK3/AKT, and NOTCH are examples of signaling pathways that are involved in the viral infection by cellular response [10]. Other processes, such as cellular miRNA "evasion" and flaws in miRNA production, may, on the other hand, favor viral replication and virus survival in host cells [11].
In this study, we investigate the novel expression of miRNA 20a and miRNA 320 and their role as a biomarker in COVID-19 infection.
Methods
Patients and controls
A total of 145 COVID-19-positive patients without any chronic disease admitted to various hospitals in Baghdad/ Iraq underwent a case–control study from the period December 2021 to February 2022, and this research was approved by the College of Science/University of Baghdad's Research Ethics Committee (Ref.: CSES/0322/0071).
The patients' COVID-19 positivity was determined by a molecular test performed on nasopharyngeal swabs. They were examined 4–7 days after admission, and a chest computerized tomography (CT) scan was done to confirm the diagnosis. Patients included those who had a positive molecular test, and a COVID-19-positive CT scan were critically unwell in the intensive care unit, and had low oxygen saturation (SPO2 < 93). Healthy control samples of 145 healthy individuals were also included. They were blood donors (Iraqi National Blood Bank), and their serum profile for anti-infectious pathogens antibodies tested negative. They were also tested for anti-COVID-19 IgG and IgM antibodies using the COVID-19 IgG/IgM Rapid Test Cassette (Inzek B.V., The Netherlands), and their results were negative.
Blood sampling
Each participant's 5 milliliters of whole blood had to be drawn directly from the vein into an EDTA-containing tube, and the procedure had to be carried out in an aseptic way.
miRNA extraction and analysis
miRNA was isolated from the blood of both groups (patients and controls) according to [12] using the EasyPure® Blood Genomic miRNA Kit (Transgen, China) with catalogue number (ER601-01).
Primer preparation for miRNA-20a and miRNA-320 gene expression
The cDNA sequences of the housekeeping genes miRNA-U6 and miRNA-20a, as well as miRNA-320, were designed by the National Centre for Biotechnology Information (NCBI) Gene Bank database. After lyophilized primers were dissolved in nuclease-free water according to the manufacturer's instructions to form a stock solution with a concentration of 100 µl for each primer and kept at (-20°C). By mixing 10 µl of primer stock solution with 90 µl of nuclease-free water, a working solution was generated which was 10µM. The primers utilized in this investigation, as well as their related sequences, are shown in Table 1.
Gene expression
cDNA synthesis from miRNA
To evaluate the synthesis of the cDNA, the EasyScript® One-Step gDNA removal with cDNA Synthesis SuperMix method was utilized (Transgen, China) which was employed to evaluate miRNA-20a and miRNA-320 expression. Reverse transcription reactions need to be put together in an environment free of RNase. All materials were thawed, the solutions were gently mixed, and the miRNA templates were added. 20µl of components reaction were used which contained 1µl of Anchored Oligo(dT)8 1 µl Primer(0.5 µg/µl), 1µl of Random Primer(0.1 µg/µl), 10 µl of 2 × EX Reaction Mix, 1 µl of Easy Script®RT/RI Enzyme Mix, 1 µl of gDNA Remover, 1 µl RNase-free Water and 5 µl of total miRNA The thermal cycler program of cDNA was composed of three steps: Step one was for a random primer with 25°C and 10 min, step 2 for anchored oligo(dt)8 with 42°C and 15min and step 3 for inactivating reverse transcriptase enzyme with 85 oC and 5 s. The synthesized cDNA was immediately utilized as a template for PCR or stored at -20°C for lengthy periods.
Quantitative real-time PCR (qRT–PCR):
MiRNA-20a and miRNA-320 expression levels were determined using qRT-PCR. qRT-PCR SYBR Green test (TransStart® Top Green qPCR SuperMix for gene expression) was performed to validate the target miRNA expression. This operation was done depending on the manufacturer's instructions (Transgene, China). The Cepheid Real-time PCR System (Smart Cycler Technologies) and qPCR Soft software were used to run qRT-PCR. By measuring the threshold cycle, the fold change and gene expression levels were calculated (Ct), and the threshold cycle (Ct) was measured to calculate the gene expression levels and fold change. Each reaction was carried out twice. The thermal profile was used to train the cycling process for optimal cycles. One cycle of initial denaturation at 94°C for 30 s, 40 cycles for three stages of denaturation (94 oC for 5 s), annealing (56 oC for both miRNA with 10 s), and extension (72 oC for 20 s). For this gene expression, a single melting curve cycle (65–95 oC) was used for 1 min. The real-time cycler software was used to calculate the threshold cycle (Ct) for each sample. Selected gene expression data were compared to housekeeping data. The Ct technique was used for data analysis, and the results were reported as folding changes in gene expression according to Aljoubory [13].
Statistical analysis
Statistical data for the Social Sciences (SPSS version 26.0 software) was used. The results are represented by either a number and a percentage, a mean and a standard deviation (SD). To determine the significance of the differences, an independent T-test and Mann–Whitney U test were utilized. A statistically significant value was defined as one with a P-value less than 0.05.
Results
All of the studied participants were of Iraqi ethnicity. The selected clinical characteristics of the included population were no variations in demographic characteristics between the COVID-19 cases and healthy controls. The COVID-19 group consisted of 145 patients (median age at diagnosis 40.56 ± 10.19 years), who were selected from different hospitals in Baghdad/ Iraq. No significant differences in age and gender distribution were seen between the studied groups. The control group was age-matched to the COVID-19 group (i.e., median age 38.78 ± 10.57 years) and comprised 145 healthy subjects as shown in Table 2.
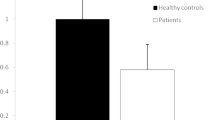
The current study examined the expression of miRNA in patients with COVID-19 since miRNAs have the potential to be biomarkers that alter post-transcriptional gene regulation. Subjects with COVID-19 had lower mean ΔCT values for miRNA-20a gene expression (4.02 ± 0.34) than controls (7.66 ± 1.11), and the observed difference was statistically significant (P-value was less than 0.0001) as shown in this investigation. This shows that the patients' miRNA-20a gene expression was considerably higher than that of the control group (Fig. 1, Table 3). The ΔCT values for miRNA-320 gene expression in COVID-19 patients (6.00 ± 0.37) were higher than that of controls (4.90 ± 0.60). Since the p-value was less than 0.0001, the observed difference was statistically significant. This shows that the miRNA-320 gene expression levels were considerably lower in the patients than in the controls (Fig. 1, Table 3).
So the fold change values for miRNA-20a gene expression in cases with COVID-19 (10.65) were higher than that of the controls (1.03) while the fold change values for miRNA-20a gene expression in cases with COVID-19 (0.48) were lower than that of the controls (1.08). This indicates that the miRNA-20a and miRNA-320 gene expression in the patients might be used as a biomarker for COVID-19 patients (Table 4).
Furthermore, receiver operating characteristic (ROC) curve analysis was used to identify miRNA-20a and miRNA-320 in blood as a biomarker for distinguishing COVID-19 patients from healthy controls. The AUC for miRNA-20a in coronavirus disease 2019 (COVID-19) patients was 1.00 (0.974 to 1.000), with optimal sensitivity and specificity values of 100% and 95%, respectively, to the cutoff value of 14.55. (Fig. 2A). The AUC for miRNA-320 in blood expression was 1.00 (0.974 to 1.000) for patients with COVID-19 compared to the control group, with optimal sensitivity and specificity values of 100% and 91%, respectively, to the cutoff value of 24.55 (Fig. 2B). As a result, the expression of miRNA-20a and miRNA-320 in blood samples has sufficient sensitivity and specificity to distinguish samples without coronavirus disease 2019 (COVID-19) from samples with COVID-19. Receiver operating characteristic (ROC) curve analysis revealed that miRNA-20a and miRNA-320 had significant sensitivity and specificity, indicating that they are suitable biomarkers for COVID-19 disease (Fig. 2).
ROC curve analysis of miRNA-20 and miRNA-320 in CΟVΙD-19 cases, miRNA-20a and miRNA-320 showed good discriminating efficiency A ROC curve of miRNA-20a (control vs. COVID-19) the AUC was 1.00 (0.974 to 1.000), the sensitivity and specificity values of 100% and 95%, respectively, the cutoff value of 14.55. B ROC curve: miRNA-320 (control vs COVID-19), the AUC was 1.00 (0.974 to 1.000), sensitivity and specificity values of 100% and 91%, respectively, the cutoff value of 24.55 AUC: area under the curve, CI: confidence interval, OR = odd ratio
Discussion
miRNAs have been identified as potential targets that could aid in the comprehension of COVID-19 infection mechanisms and the development of novel antiviral drugs [14]. In this study, the expression levels of miRNA-20a and miRNA-320 in COVID-19 patients were compared to those in healthy controls. Significantly elevated levels of miRNA-20a were observed in COVID-19 patients compared to controls. ROC curve analysis verified this result's accuracy with a P-value of 0.0001, which agreed with Liu et al. who reported that miRNA20a was up-regulated in pediatric pneumonia and human lung adenocarcinoma A549 cells [15]. Similar findings were found in investigations on other chest diseases. Xu et al. also discovered that non-small cell lung cancer (NSCLC) patients had higher levels of miRNA-20a than healthy controls [16]. However other studies showed different investigation to the findings of Giannella et al. 2022 and Li et al. 2020, it has been observed that the expression of miRNA-20a is notably reduced in individuals diagnosed with COVID-19 in comparison to the control groups [17, 18]. MIR20, miRNA-20, MIRH1, MIRN20,MIR17HG, MIRN20A, MIRHG1, miRNA20A, C13orf25, miRNA20A, and hsa-miRNA-20a or hsa-miRNA-20 are all names for human miRNA-20a. It's found on chromosome 13q31.3.MicroRNA-20a gene expression was significantly greater in COVID-19 patients compared to controls. Another research discovered that overexpression of miRNA-20a decreases the production of mediators of inflammation such as IL-2, IL-6, and IL-8 [19]. This miRNA was shown to be downregulated in multiple sclerosis (MS) patients compared with the control group [20]. However, because of the significant difference in miRNA20a gene expression between patients and controls, it might be used as a biological marker to identify COVID-19 patients. Previous research has shown that miRNA-20a is associated with cell proliferation and progression through the cell cycle by modulating the expression of transcription factor E2F1. Fan et al. [21] also reported that miRNA-20a overexpression was able to promote cell proliferation and invasion by inhibiting the expression of targeted amyloid precursor protein in ovarian cancer cells. In addition, miRNA-20 has been shown to play an essential role in inflammatory responses. Philippe et al. [22] reported that miRNA-20 inhibited the production of inflammatory cytokines such as IL-6, C-X-C motif chemokine ligand 10, IL-1, and TNF- in LPS-activated fibroblast-like synoviocytes. In contrast, it has been reported that miRNA-20a promotes macrophage inflammatory responses by modulating the expression of signal-regulatory protein [23].
The current work describes the function of miRNA-20a in COVID-19 and offers details that can be applied to future studies on the biomarker function of miRNA-20a.
In COVID-19 patients, miRNA-320 levels were significantly lower than in healthy controls. ROC curve analysis confirmed the veracity of this result with a P value of 0.0001, confirming Duecker et al.'s finding that downregulation of miRNAs-320 has been documented in patients exhibiting acute mild symptoms, and this downregulation continues to decrease progressively in patients with severe progression of the disease [24].
miRNA-320 was shown to be downregulated in patients who had COVID-19 symptoms. Deep vein thrombosis (DVT) patients have already been studied using the miRNA-320 [25,26,27]. DVT is a serious consequence that affects COVID-19 patients as well, thus it's critical to identify high-risk COVID-19 patients so that thrombosis prophylaxis can be started right away [28]. Jiang et al. discovered that miRNA-320a and miRNA-320b substantially increase concentrations of D-dimer in patients with deep vein thrombosis (DVT) but not in those with post-thrombotic syndrome contradicts our results [29]. A preventive mechanism against abrupt thrombosis in COVID-19 patients may have caused a decrease in miRNA-320 expression, which could account for these disparities. MiRNA-320 downregulation protects against cardiac ischemia–reperfusion injury by promoting Nrf2 expression, according to Zhu et al.[25].
Downregulation of miRNA-320 has been related to Nrf2's control of oxidative stress and inflammation [26]. Along with miRNA-320, many miRNAs were discovered to be dysregulated in the patient groups. The two microRNAs with the greatest levels, miRNA-374a-3p and miRNA-15a-3p, have been associated with acute nephritis and neuroinflammation, respectively. A further molecular diagnostic miRNA biomarker for liver fibrosis in people with chronic hepatitis B is miRNA-4721, which has been proposed. The potential diagnostic biomarker for biliary atresia, Hsa-miRNA-4429, has been postulated [30,31,32,33,34].
Even though little to no study has been conducted on these miRNAs, it is vital to keep them in mind as possible indications of the devastating COVID-19 illness. Earlier COVID-19 research focused on miRNAs such as 126-5p, miR-27, miR-21-5p, miR-146, and miR-142 [35, 36]. Garg et al. [36] discovered dysregulated levels of miR-21-5p and miR-126-5p, two miRNAs linked to inflammatory processes, in the circulating microRNAs of seriously ill COVID-19 patients. When De Gonzalo-Calvo et al. [37], compared the profiles of circulating miRNA of patients who did not require critical care to those admitted to the intensive care unit, they discovered an elevation of miRNA-27a-3p, miRNA-148a-5p, and miRNA-27b-3p that distinguished ICU from non-ICU patients[37]. Those who investigated cellular miRNAs during the acute and post-acute phases of COVID-19 discovered that severe patients had higher levels of miRNA-29a-3p and miRNA-146a-3p [38]. The value of miRNAs as circulating diagnostic biomarkers was determined using the AUC of the ROC curves. According to Hosmer and Lemeshow, "0.5 = no discrimination; 0.5–0.7 = poor discrimination; 0.7–0.8 = acceptable discrimination; 0.8–0.9 = excellent discrimination; and > 0.9 = outstanding discrimination"[39]. The current research discovered that miRNA-20a in blood samples has excellent discrimination for use as a diagnostic biomarker, Other studies with the same goal found similar results with another miRNA based on the AUC of the ROC curve in the healthy group to COVID-19 patients. With AUCs of 0.959 (95% CI = 0.917–1.000, p 0.0001), Markou et al. determined that miRNA-210 was a useful biomarker for distinguishing patients from healthy individuals using ROC curve analysis [40]. The findings of the numerous studies frequently differ, sometimes noticeably, in terms of the sample preparation, materials testing, and molecular approach, as well as the severity of the patients, evaluated.
Conclusion
The results of our investigation indicate a higher frequency of expression for miRNA-20a, whereas miRNA-320 exhibited a lower frequency of expression.This finding implies that miRNA-20a and miRNA-320 have the potential to function as early indicators for the identification of individuals with COVID-19. ROC analysis was employed to assess the specificity and sensitivity of these miRNAs in COVID-19 patients.
Availability of data and materials
On request, the corresponding author will provide the data that back up the study's conclusions.
Abbreviations
- CoVs:
-
Coronaviruses
- ORF:
-
Open reading frame
- NSPs:
-
Non-structural proteins
- TMPRSS2:
-
Transmembrane serine protease 2
- qRT–PCR:
-
Quantitative real-time PCR
- ROC:
-
Receiver operating characteristic
References
Demirci S, Adan A (2020) Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. Peer J 5(8):e9369. https://doi.org/10.7717/peerj.9369
Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH et al (2005) Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79(2):884–895. https://doi.org/10.1128/JVI.79.2.884-895.2005
Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, Liu W, Bi Y, Gao GF (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24(6):490–502. https://doi.org/10.1016/j.tim.2016.03.003
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
WHO Coronavirus Disease (COVID-19) Dashboard. 2020. https://covid19.who.int/
Rad AH, McLellan AD (2020) Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. Int J Mol Sci 21(13):4807. https://doi.org/10.3390/ijms21134807
Canatan D, Sanctis VD (2020) The impact of microRNAs (miRNAs) on the genotype of coronaviruses. Acta Biomed 91(2):195–198. https://doi.org/10.23750/abm.v91i2.9534
Bernier A, Sagan SM (2018) The diverse roles of microRNAs at the Host-Virus interface. Viruses 10(8):440. https://doi.org/10.3390/v10080440
Girardi E, López P, Pfeffer S (2018) On the importance of host MicroRNAs during viral infection. Front Genet 9:439. https://doi.org/10.3389/fgene.2018.00439
Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A et al (2016) A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe 19(3):409–423. https://doi.org/10.3389/fgene.2018.00439
Trobaugh DW, Klimstra WB (2017) MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol Med 23(1):80–93. https://doi.org/10.1016/j.molmed.2016.11.003
Mohamed HA, Abdelkafy AE, Khairy RMM et al (2023) MicroRNAs and cytokines as potential predictive biomarkers for COVID-19 disease progression. Sci Rep 13:3531. https://doi.org/10.1038/s41598-023-30474-6
Aljoubory HM, Altaee MF (2021) Correlation study between three different genes expression and chronic myeloid leukaemia in Iraq. Iraqi J Agric Sci 52(3):611–619
Fani M, Zandi M, Ebrahimi S, Soltani S, Abbasi S (2021) The role of miRNAs in COVID-19 disease. Futur Virol 16(4):301–306. https://doi.org/10.2217/fvl-2020-0389
Liu Z, Yu H, Guo Q (2018) MicroRNA-20a promotes inflammation via the nuclear factor-κB signaling pathway in pediatric pneumonia. Mol Med Rep 17(1):612–617
Xu X, Zhu S, Tao Z, Ye S (2018) High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Med 7(1):21–31. https://doi.org/10.1002/cam4.1238
Giannella A, Riccetti S, Sinigaglia A, Piubelli C, Razzaboni E, Di Battista P, Agostini M, Dal Molin E, Manganelli R, Gobbi F, Ceolotto G, Barzon L (2022) Circulating microRNA signatures associated with disease severity and outcome in COVID-19patients. Front Immunol 11(13):968991. https://doi.org/10.3389/fimmu.2022.968991
Li C, Hu X, Li L, Li JH (2020) Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal 34(10):e23590. https://doi.org/10.1002/jcla.23590
Barbu MG, Condrat CE, Thompson DC, Bugnar L, Cretoiu D, Toader OD, Suciu N, Voinea SC (2020) MicroRNA involvement in signaling pathways during viral infection. Front Cell Dev Biol 8:143. https://doi.org/10.3389/fcell.2020.00143
Abdullah D, Aloubaidy MR (2022) Genetic polymorphism of caspase8 and 9 in Iraq. Iraqi J Agric Sci 53(3):505–514. https://doi.org/10.36103/ijas.v53i3.1558
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T, Liu M, Li X, Tang H (2010) miR-20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells. Acta Biochim Biophys Sin (Shanghai) 42:318–324. https://doi.org/10.1093/abbs/gmq026
Philippe L, Alsaleh G, Pichot A, Ostermann E, Zuber G, Frisch B, Sibilia J, Pfeffer S, Bahram S, Wachsmann D, Georgel P (2013) MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann Rheum Dis 72:1071–1079. https://doi.org/10.1136/annrheumdis-2012-201654
Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L, Zhang J, Li D, Gu H, Zhang CY et al (2013) MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein α. J Allergy Clin Immunol 132:426–436. https://doi.org/10.1016/j.jaci.2013.02.005
Duecker RP, Adam EH, Wirtz S, Gronau L, Khodamoradi Y, Eberhardt FJ, Donath H, Gutmann D, Vehreschild MJGT, Zacharowski K, Kreyenberg H, Chiocchetti AG, Zielen S, Schubert R (2021) The MiR-320 family is strongly downregulated in patients with COVID-19 induced severe respiratory failure. Int J Mol Sci 22(19):10351. https://doi.org/10.3390/ijms221910351
Zhu SY, Wu QY, Zhang CX, Wang Q, Ling J, Huang XT et al (2018) miR-20a inhibits the killing effect of natural killer cells to cervical cancer cells by downregulating RUNX1. Biochem Biophys Res Commun 505(1):309–316. https://doi.org/10.1016/j.bbrc.2018.09.102
Li S, Qiang Q, Shan H, Shi M, Gan G, Ma F, Chen B (2016) MiR-20a and miR-20b negatively regulate autophagy by targeting RB1CC1/FIP200 in breast cancer cells. Life Sci 147:143–152. https://doi.org/10.1016/j.lfs.2016.01.044
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L (2013) Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J Exp Clin Cancer Res 32:21. https://doi.org/10.1186/1756-9966-32-21
Bai X, Han G, Liu Y, Jiang H, He Q (2018) MiRNA-20a-5p promotes the growth of triple-negative breast cancer cells through targeting RUNX3. Biomed Pharmacother 103:1482–1489. https://doi.org/10.1016/j.biopha.2018.04.165
Jiang Z, Ma J, Wang Q, Wu F, Ping J, Ming L (2018) Combination of circulating miRNA-320a/b and D-Dimer improves diagnostic accuracy in deep vein thrombosis patients. Med Sci Monit 24:2031–2037. https://doi.org/10.12659/msm.906596
Starikova I, Jamaly S, Sorrentino A, Blondal T, Latysheva N, Sovershaev M, Hansen JB (2020) Role of microRNAs in Venous Thromboembolism. Int J Mol Sci 21:2602. https://doi.org/10.3390/ijms21072602
Cai C, Guo Y, You Y, Hu K, Cai F, Xie M, Yang L, Ling K, Ye D, Misra et al. Deep venous thrombosis in COVID-19 patients: a cohort analysis. Clin Appl Thromb Hemost. 26: 1076029620982669. doi: https://doi.org/10.1177/1076029620982669.
Zhu XA, Gao LF, Zhang ZG, Xiang DK (2019) Down-regulation of miR-320 exerts protective effects on myocardial I-R injury via facilitating Nrf2 expression. Eur Rev Med Pharmacol Sci 23(4):1730–1741. https://doi.org/10.26355/eurrev_201902_17135
Wang M, Wei J, Shang F (2021) Down-regulation of lncRNA SNHG5 relieves sepsis-induced acute kidney injury by regulating the miR-374a-3p/TLR4/NF-κB pathway. J Biochem 169(5):575–583. https://doi.org/10.1093/jb/mvab008
Dong R, Shen Z, Zheng C, Chen G, Zheng S (2016) Serum microRNA microarray analysis identifies miR-4429 and miR-4689 are potential diagnostic biomarkers for biliary atresia. Sci Rep 6:21084. https://doi.org/10.1038/srep21084
Lu L, Dai WZ, Zhu XC, Ma T (2021) Analysis of serum miRNAs in Alzheimer’s disease. Am J Alzheimers Dis Other Demen 36:15333175211021712. https://doi.org/10.1177/15333175211021712
Garg A, Seeliger B, Derda AA, Xiao K, Gietz A, Scherf K, Sonnenschein K et al (2021) Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur J Heart Fail 23(3):468–475. https://doi.org/10.1002/ejhf.2096
De Gonzalo-Calvo D, Benítez ID, Pinilla L, Carratalá A, Moncusí-Moix A, Gort-Paniello C et al (2021) Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl Res 236:147–159. https://doi.org/10.1016/j.trsl.2021.05.004
Donyavi T, Bokharaei-Salim F, Baghi HB, Khanaliha K, Janat-Makan MA, Karimi B et al (2021) Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a–3p, 155–5p, and let-7b-3p in PBMC. Int Immunopharmacol 97:107641. https://doi.org/10.1016/j.intimp.2021.107641
Hosmer DW, Lemeshow S , Sturdivant RX. Applied Logistic Regression, 2013. 3rd ed.; Wiley: Hoboken, NJ, USA, 177. doi:https://doi.org/10.1002/9781118548387
Markou A, Zavridou M, Sourvinou I, Yousef G, Kounelis S, Malamos N et al (2016) Direct comparison of metastasis-related mirnas expression levels in circulating tumor cells, corresponding plasma, and primary tumors of breast cancer patients. Clin Chem 62(7):1002–1011. https://doi.org/10.1373/clinchem.2015.253716
Acknowledgements
No applicable
Funding
This investigation was self-funded. No financial assistance.
Author information
Authors and Affiliations
Contributions
RMA, HWA, and LAY made significant contributions to the preparation of this study through the collection of samples from patients and healthy controls, the design of the work protocol, participation in the practical portion of the study, and writing and editing of the manuscript. The concept development, analysis, and interpretation, as well as the writing and revision of the final manuscript, were all completed by all authors, who also read and approved the document.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
The study received ethics approval and participant consent because it made use of pre-existing COVID-19 case-based data and contextual data from routine COVID-19 epidemiological reports and humanitarian situation reports. The national COVID-19 incident manager gave administrative approval for the study, which is categorized as operational research and is done in order to support an evidence-based pandemic response. The research was also approved by the College of Science/University of Baghdad's Research Ethics Committee (Ref.: CSES/0322/0071).
Consent for publication
The patient's permission to publish had been obtained.
Competing interests
This investigation contains no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abed, R.M., Abdulmalek, H.W. & Yaaqoob, L.A. The role of miRNA20a and miRNA320 in Iraqi patients with COVID-19: a case–control study. Egypt J Med Hum Genet 24, 68 (2023). https://doi.org/10.1186/s43042-023-00451-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-023-00451-9