Abstract
Background and aim
The primary factor in sudden cardiac death is coronary artery disease. We intended to discover the diagnostic worth of circulating tumor necrosis factor like cytokine 1A (TL1A) and free fatty acid receptor 2 (FFAR2) as early, noninvasive indicators for individuals with stable angina (SA) and unstable angina (UA).
Methods
In all, 90 people were enrolled in the current case–control study: 30 patients with SA, 30 patients with UA, and 30 healthy volunteers. Circulating TL1A and FFAR2 gene expression levels were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). FBG, TC, TG, and HDL-C were assessed by spectrophotometry, while hs-CRP and troponin T were measured by ELISA.
Results
Circulating TL1A expression was significantly elevated in SA (P < 0.001) and UA patients (P < 0.001) as compared to controls and also was significantly higher in UA patients (P < 0.001) as compared to SA patients. Circulating FFAR2 expression was significantly decreased in SA (P < 0.001) and UA patients (P < 0.001) in comparison with controls and was significantly lowered in UA patients (P = 0.001) in comparison with SA patients. Our results show that TL1A and FFAR2 were sensitive and specific biomarkers for discriminating SA patients from controls. Moreover, TL1A and FFAR2 displayed a remarkable ability to distinguish UA from SA. Multivariate regression analysis revealed that TL1A, FFAR2, FBG, TC, TG, LDL-C, and Troponin T were independent risk factors for SA, while TL1A, TG, and hs-CRP were independent risk factors for UA. TL1A has a significant positive correlation with LDL-C (r = 0.406, P = 0.001), hs-CRP (r = 0.673, P < 0.001), and troponin T (r = 0.653, P < 0.001). There was a significant inverse relationship between FFAR2 and each of TL1A (r = − 0.858, P < 0.001), FBG (r = − 0.325, P = 0.011), TC(r = − 0.306, P = 0.017), TG (r = − 0.368, P = 0.004), LDL-C (r = − 0.413, P = 0.001), hs-CRP (r = − 0.737, P < 0.001), and troponin T (r = − 0.715, P < 0.001).
Conclusion
Gene expression of TL1A and FFAR2 is a good new blood-based molecular indicator for early detection of SA and UA. Early detection of a possible UA is crucial for initiating appropriate treatment that results in better patient health.
Similar content being viewed by others
Introduction
Blood flow to the myocardium is reduced by disorders known as coronary artery disease (CAD). The primary cause of blood flow limitation is coronary atherosclerosis, which results in plaque development. Acute coronary syndrome (ACS) might be brought on by destabilization and subsequent plaque rupture [1]. CAD is a significant issue for public health, and it continues to be the main factor in sudden cardiac death worldwide. [2]. Two clinical types of CAD are SA and ACS, which includes UA and acute myocardial infarction (AMI) [3]. The higher risk of death is tightly linked to ACS [4].
Physicians must perform early clinical identification and accurate CAD diagnosis to begin appropriate therapy and, as a result, avert unexpected cardiac death. Coronary angiography, however, the most effective invasive method for identifying CAD patients, has certain disadvantages, such as a high price, radiation risks, and restricted accessibility. Therefore, there is a clinical necessity to identify novel biomarkers found in the blood for CAD early detection [2].
The tumor necrosis factor (TNF) superfamily has a new subgroup called TL1A. TL1A affects immune cell proliferation, activation, and differentiation in a pleiotropic manner. It not only induces apoptosis and activates nuclear factor kappa B (NF-kB), but it also takes part in lymphocyte activation and proliferation, control of immune and inflammatory responses in the body, and the development and advancement of atherosclerosis and other disorders. TL1A plays a role in atherosclerosis by producing pro-inflammatory cytokines and chemokines, as well as reducing plaque stability by activating enzymes that break down the extracellular matrix [5].
Endothelial cells largely produce TLA1 in response to stimulation with TNF-α and Interleukin-1 (IL-1). TL1A levels are elevated in ulcerative colitis patients, and ankylosing spondylitis, primary biliary cirrhosis, and autoimmune rheumatic illness. It was recently discovered that TL1A has a role in the inflammatory process of coronary atherosclerosis [6].
FFAR2 has another name; G protein-coupled receptor 43 (GPR43). It is found on chromosome 19q13.1 among a group of typical intron-free genes. FFAR2 belongs to G protein-coupled receptor class A. Intestinal bacteria create short-chain fatty acids in significant part by fermenting undigested carbohydrates and dietary fibers, which further activate FFAR2. Seven hydrophobic areas are found in the FFAR2 protein, which corresponds to the transmembrane helix. As a signaling molecule, FFAR2 is involved in blood glucose, inflammation, and serum lipid regulation. Inflammation and elevated levels of glucose and cholesterol in the blood raise the possibility that healthy persons may be affected by AMI [7].
In the current study, we assessed TL1A and FFAR2 mRNA degrees of expression in SA and UA patients circulating blood. We also tried to evaluate the possibility of the use of TL1A and FFAR2 as potential biomarkers for the early detection of SA and UA. Furthermore, we studied the relationship between TL1A, FFAR2, and some laboratory parameters to learn more about the pathophysiology of SA and UA.
Patients and methods
Study subjects
Case–control research was used in the current study. It was conducted on 60 patients who attended the cardiac catheter unit, Cardiology Department, Faculty of Medicine, Assiut University, during the period from January 2021 to July 2021. They were categorized into 30 patients with SA, and 30 patients with UA. Patient features were anonymously analyzed in addition to 30 unrelated healthy controls who were admitted to the hospital with chest pain, normal cardiac enzyme levels, coronary angiography, and an ECG.
According to the guidelines provided by ACC/AHA, At least one major epicardial artery must have a stenosis of 50% or higher for there to be CAD, as determined by coronary angiography, CAD was also classified as stable or unstable [8, 9]. Exclusion criteria for the study comprised patients with recent AMI, congenital cardiac disease, cardiomyopathy, heart failure, hepatitis, hepatic failure, renal failure, blood disorders, malignancy, and autoimmune illnesses as arthritis and inflammatory bowel disease. Subjects’ demographics, full history taking, and relevant comorbidities were obtained and recorded.
Blood sample collection
After an overnight fast, a total of 5 ml of antecubital venous blood was obtained from the participants. Two milliliters of whole blood was placed in EDTA tubes and maintained at − 80 °C until qRT-PCR analysis of TL1A and FFAR2, whereas 3 ml of blood was placed in plain tubes and allowed to coagulate. The serum was obtained by centrifuging the tubes at 3000 rpm for 20 min at 4 °C and then storing them at − 20 °C for further analysis of fasting blood glucose (FBG), lipid profile, hs-CRP, and troponin T.
Biochemical analyses
Spectrophotometric techniques were used to determine serum levels of FBG, total cholesterol (TC), triglycerides (TG), and high-density lipoprotein cholesterol (HDL-C) using commercially available colorimetric assay kits (catalog no. GL1320, CH1220, TR2030, and CH1230, respectively, provided by Biodiagnostic, Egypt) based on the directions given by the manufacturer. The Friedewald formula was used to compute low-density lipoprotein cholesterol (LDL-C):
Human ELISA Kits supplied by OriGene Technologies, Inc. USA catalog no. EA101010 and RayBiotech, Inc. USA catalog no. ELH-Troponin T, respectively, were used to estimate the level of hs-CRP and Troponin T in the blood using the ELISA technique by doing what the manufacturer instructs.
Genetic analysis
Total RNA was extracted from whole blood using Thermo Scientific GeneJET Whole Blood RNA Purification Mini Kit (Catalog number: K0761). For cDNA reverse transcriptase reactions, Thermo Scientific RevertAid First Strand cDNA Synthesis Kit Catalog number: K1622 was utilized. The following conditions were used to PCR amplify cDNA in a thermal cycler:
40 cycles of denaturation for 10 s at 95 °C, annealing for 30 s at 56 °C, and an extension step for 30 s at 72 °C were performed after the initial denaturation for 2 min at 95 °C. Applied Biosystem Step OnePlus™ software was used to perform real-time PCR. The reaction uses Thermo Fisher Scientific's PowerUp™ SYBR™ Green Master Mix, (Catalog number: A25742). The expression of each gene was normalized to GAPDH as a control and expressed as a fold change. Using the comparative 2−ΔΔCT approach, relative mRNA expression levels of all investigated genes were calculated [10].
Primer sequence TL1A, FFAR2 and GAPDH were:
-
TL1A F: 5′-AAG GAC AGG AGT TTGCAC CTTCA-3′
-
; R: 5′-AAG TGC TGT GTG GGA GTT TGTCT-3′
-
FFAR2: F: 5′CTTCGGACCTTACAACGTGTC3′
-
; R: 3′CTGAACACCACGCTATTGAC5′
-
GAPDH: F: 5′TGTGGGCATCAATGGATTTGG3′
-
; R 3′ACACCATGTATTCCGGGTCAAT5′
Statistical analysis
Continuous data were presented as mean ± SD, while categorical variables were presented as a number (%). The chi-square (χ2) test was used to evaluate categorical data. Based on the normality of the data, the Student’s t test and Mann–Whitney test were used. A one-way analysis of variance (ANOVA) was used to examine the comparability between the groups. Multivariate regression analysis was performed to determine the most significant predictors for the stable and unstable angina. Correlations were examined using Pearson's correlation coefficient test. Statistical significance was defined as P < 0.05. IBM SPSS software version 20 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7 Software (San Diego, California, USA) were used to conduct all statistical analyses. MedCalc software was used to compute the ROC curve and cutoff value.
Results
Clinical characteristics of the study population
The baseline characteristics of the three groups are presented in Table 1. The SA group (n = 30) had 20 males and 10 females (49.37 ± 4.97 years). The UA group (n = 30) consisted of 23 men and 7 women with 47.23 ± 5.49 years average age. The control group (n = 30) had an average age of 48.5 ± 5.06 years and was made up of 19 males and 11 women. Age, gender, BMI, diabetes, hypertension, or smoking did not display significant differences across the three groups (P > 0.05). FBG, TC, TG, LDL-C, hs-CRP, and troponin T levels were significantly higher in SA patients than in controls, as well as in UA patients than in SA patients. The HDL-C of the SA patients, on the other hand, was significantly lower than that of the controls and in UA was lower than that of SA. Table 1 displays all of the data in detail.
Gene expression pattern of circulating TL1A and FFAR2
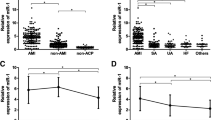
Fold change gene expression of TL1A and FFAR2 in the three studied groups is presented in Fig. 1. When comparing SA and UA patients to controls, there was a significant increase in TL1A fold change gene expression (P < 0.001). Furthermore, the level of TL1A expression in UA patients was significantly higher than in SA patients (P < 0.001). In terms of FFAR2 fold change gene expression, compared to controls, SA and UA patients both had considerably lower levels. (P < 0.001). Furthermore, it was significantly lower in UA patients than in SA patients (P = 0.001).
Fold change gene expression (2^-ΔΔCt) of TL1A and FFAR2 in SA and UA patients and controls. The data are presented as the mean ± SEM. FFAR2 free fatty acid receptor 2, SA stable angina, SEM Standard error of mean, TL1A tumor necrosis factor like cytokine 1A, UA unstable angina, and P value < 0.05 is considered statistically significant
Diagnostic significance of circulating TL1A and FFAR2 gene expression in SA and UA patients.
A receiver operating characteristic (ROC) curve analysis was used to explore the diagnostic accuracy of circulatory TL1A and FFAR2 as possible biomarkers of SA and UA. With an area under the curve (AUC) of 0.971 and 0.891, cutoff > 2.3762 and < 0.6885, sensitivity 96.67% and 96.67%, specificity 93.33% and 86.67%, respectively, the ROC curves of TL1A and FFAR revealed a considerable separation between SA patients and controls (Fig. 2A). Similarly, with AUCs of 0.949 and 0.987, cutoffs > 26.946 and < 0.0169, sensitivity 90% and 93.33%, and specificity 96.67% and 96.67%, respectively, the ROC curves of TL1A and FFAR2 indicated a significant capacity to differentiate between SA and UA patients (Fig. 2B). These findings suggested that circulating TL1A and FFAR2 could be employed as potential early diagnostic biomarkers for SA and UA.
Multivariate regression analysis: predictors of SA and UA
Multivariate regression analysis revealed that TL1A, FFAR2, FBG, TC, TG, LDL-C, and Troponin T are significant predictors for stable angina while TL1A, TG, and hs-CRP are significant predictors for unstable angina (Table 2).
Correlation between TL1A and FFAR2 with clinical characteristics in SA and UA patients
We also investigated the correlations of gene expression levels of TL1A and FFAR2 with clinical characteristics in patients with SA and UA. TL1A has a significant positive correlation with LDL-C (r = 0.406, P = 0.001), hs-CRP (r = 0.673, P < 0.001), and troponin T (r = 0.653, P < 0.001), according to our findings (Fig. 3B–D). There was a significant inverse relationship between FFAR2 and each of TL1A (r = − 0.858, P < 0.001), FBG (r = − 0.325, P = 0.011), TC (r = − 0.306, P = 0.017), TG (r = − 0.368, P = 0.004), LDL-C (r = − 0.413, P = 0.001), Hs-CRP (r = − 0.737, P < 0.001) and troponin T (r = − 0.715, P < 0.001); (Fig. 3A, E–J).
Correlation between: A TL1A and FFAR2; B TL1A and LDL-C; C TL1A and hs-CRP; D TL1A and troponin T; E FFAR2 and FBG; F FFAR2 and TC; G FFAR2 and TG; H FFAR2 and LDL-C; I FFAR2 and hs-CRP; J FFAR2 and troponin T among 30 SA and 30 UA patients. Significant positive correlation was found between TL1A and each of B LDL-C (r = 0.406, P = 0.001); C hs-CRP (r = 0.673, P < 0.001) and D troponin T (r = 0.653, P < 0.001). A significant negative correlation was found between FFAR2 and each of A TL1A (r = − 0.858, P < 0.001); E FBG (r = − 0.325, P = 0.011); F TC (r = − 0.306, P = 0.017); G TG (r = − 0.368, P = 0.004); H LDL-C (r = − 0.413, P = 0.001); I hs-CRP (r = − 0.737, P < 0.001) and J troponin T (r = − 0.715, P < 0.001). FBG fasting blood glucose, FFAR2 free fatty acid receptor 2, hs-CRP high-sensitivity C-reactive protein, LDL-C low-density lipoprotein cholesterol, r correlation coefficient, TC total cholesterol, TG triglycerides, TL1A tumor necrosis factor like cytokine 1A, and P value < 0.05 is considered statistically significant
Discussion
CAD is a chronic inflammatory condition that progresses from chest discomfort to SA and then to severe UA or AMI [11]. Early and precise diagnosis is critical for effective intervention to improve disease prognosis and reduce the risk of AMI [12]. To the best of our knowledge, the present study is the first study that evaluated the circulating TL1A and FFAR2 gene expression in SA and UA patients.
The level of circulating TL1A gene expression was evaluated in this study. In comparison with controls, it was significantly upregulated in SA and UA patients. It was also significantly elevated in UA patients than in SA patients. By obtaining AUC values from the ROC curve, we investigated the diagnostic potential of circulating TL1A gene expression levels. With an AUC of 0.971, 96.67% sensitivity, and 93.33% specificity, we found that TL1A can distinguish SA from controls. We also found that TL1A has the ability to distinguish SA from UA, with an AUC of 0.949, 90% sensitivity, and 96.67% specificity. Furthermore, our results revealed that TL1A significantly predicts SA and UA.
Unstable angina is typically caused by thrombotic reactions triggered by the rupture of unstable plaque [13]. A fibrous cap composed of vascular smooth muscle cells and extracellular matrix molecules keeps atherosclerotic plaques stable. TL1A, on the other hand, can promote the synthesis of matrix metalloproteinases 1, 9, and 13 in monocytes by interacting with DR3 [14]. The fibrous cap may then be degraded by these matrix metalloproteinases, resulting in plaque rupture and, as a result, acute symptoms [15]. This mechanism may explain the upregulation of TL1A in SA and UA.
Our findings are consistent with those of Chen et al. [5] who revealed that levels of TL1A in the UA group's coronary and peripheral blood and plasma were greater than those in the control group. Also, the present study goes hand in hand with a study conducted by Li et al. [15] who indicated the high sensitivity and specificity of TL1A and the possibility that it may be a potentially valuable biomarker for diagnosing CAD. Endothelial cells produce the majority of the TLA1 gene in response to TNF-α and IL-1 stimulation. DR3 is a soluble cell surface receptor that regulates apoptosis, promotes actin rearrangement, and causes endothelial cells to release intercellular adhesion molecule 1, vascular cell adhesion molecule 1, and interleukin 8. TL1A activates DR3, which activates several inflammatory pathways.
The level of TL1A has previously been used to assess severity and prognosis in CAD patients who require coronary artery bypass grafting. TL1A and DR3 plasma levels were considerably higher in patients with CAD compared to controls, suggesting that they play a role in CAD development [15].
The current study measured hs-CRP and troponin T, the gold standards for detecting CAD, in patients with SA and UA. Lipid profile and FBG levels were also assayed, as well as correlated with TL1A. Significant positive correlations between TL1A and hs-CRP, troponin T, and LDL-C were found. These findings were in line with those of Akyüz et al. [6] who discovered a correlation between hs-CRP and TL1A levels, suggesting that raised TL1A levels in CAD may have a pro-inflammatory effect comparable to increased CRP levels. The fact that TL1A levels were an independent CAD predictor shows that TL1A's role in inflammation is complementary to that of hs-CRP.
In agreement, Stamatelopoulos et al. investigated the role of TL1A in the development of atheromatous plaques and found that it can be utilized to diagnose and predict prognosis in individuals with stable CAD and ACS. This could explain why TL1A plays a pathogenic role in atherosclerosis and plaque disruption [16]. Anti-TNF monoclonal antibodies or the next generation of anti-TNF drugs that block TL1A-mediated interactions may have therapeutic significance in cardiovascular disease [17]. Additionally, Chen et al. [5] noticed that coronary TL1A levels were substantially linked with peripheral TL1A levels as well as TL1A levels in coronary thrombus and local plaques. The fact that TL1A has been associated with coronary thrombus burden, inadequate coronary flow/no coronary reflow, and these conditions suggests that it plays a role in both the rupture of small atherosclerotic plaques and the thrombosis of the coronary artery.
This underpins the significance of the biomarker in the diagnosis of SA patients. Its increased level of expression in patients with UA may also be a potential indicator of a bad prognosis.
Short-chain fatty acids play a role in the cellular metabolism of fatty acids, glucose, and cholesterol in a variety of peripheral tissues, both directly and genetically. Because of their immunoregulatory properties and impact on the metabolism of lipids, cholesterol, and glucose, short-chain fatty acids may help to create metabolic conditions that support the preservation or restoration of endothelial function, thereby reducing the risk of cardiovascular disease onset or progression [18].
Free fatty acid receptor 2 (FFAR2) is a member of G protein-coupled receptors [19]. Enteroendocrine, pancreatic β, Adipocytes, and different inflammatory cells such as neutrophils and macrophages all express FFAR2. FFAR2 expression in the liver has not been reported. Short-chain fatty acids acetate and propionate, followed by butyrate, are the major gut microbial metabolites that FFAR2 selectively binds [20]. This provides a method by which the gut microbiota can influence various physiological processes in multiple cells. FFARs have become prospective targets in many pathophysiological circumstances as a metabolic modulator of various metabolic diseases due to their extensive distribution and activity as a signaling molecule involved in regulating multiple metabolic homeostatic systems in the human body [21].
The present study looked into FFAR2 gene expression and revealed a significant reduction in FFAR2 fold change gene expression in both SA and UA patients when compared to controls, with UA patients having significantly lower expression than SA patients. The ROC curve for FFAR2 revealed that at AUC (0.891), it had 96.67% sensitivity and 86.67% specificity, implying that it might be used as a biomarker for early diagnosis of SA. We also found that FFAR2 has can discriminate SA from UA, with an AUC of 0.987, 93.33% sensitivity, and 96.67% specificity.
In support of our results, Ruan et al. [7] found that at both the gene and protein levels, Patients with AMI had lower levels of FFAR2 gene expression in their peripheral blood than those in the control group. Through its impact on hormone production and inflammation, FFAR2 affects lipid metabolism and glucose levels [20]. AMI risk is increased in healthy persons with inflammation and increased blood glucose, and cholesterol levels. By modifying the pathways indicated above, low FFAR2 expression could be used as a biomarker for predicting the occurrence of AMI [7].
Both Gi/o and Gq/11 proteins are known to link to FFAR2 [22]. Gq protein signaling increases intracellular calcium and encourages the activation of the MAPK cascade, whereas signaling via Gi/o proteins limits cAMP synthesis and activates the ERK cascade [23]. The pro-inflammatory transcription factor nuclear factor NFk-B is inhibited by FFAR2's activation of -arrestin2, which also reduces the manufacturing of pro-inflammatory cytokines like interleukin-1β and IL-6 [24]. FFAR2 is linked to leukocyte migration and cytokine secretion, suggesting that short-chain fatty acids' anti-inflammatory effects are mediated via FFAR2 [25].
The current study found a significant negative correlation between FFAR2 gene expression levels and other examined biomarkers, including TL1A, FBG, TC, TG, LDL-C, hs-CRP, and troponin T. The relatively low expression of the FFAR2 gene, which increases FBG, TC, TG, and LDL-C levels, is one of the mechanisms by which low FFAR2 promotes SA and UA.
The current correlation in this study indicates FFAR2's position as an early diagnostic indicator in patients with SA while being a poor prognostic marker in patients with UA. FFAR2 signaling modulates the production of peptide tyrosine tyrosine (PYY) and glucagon-like peptide 1 (GLP-1) and may open the way for FFAR2 to be considered as a therapeutic target for diabetes, as GLP-1 increases are advantageous in blood glucose regulation [26]. FFAR2 is involved in the control of appetite and insulin signaling in the intestines [27]. In animal model experiments, acetate, propionate, and butyrate increase the uptake of cholesterol from the circulation by the liver and thus decrease plasma cholesterol. Propionate is also a powerful inhibitor of cholesterol synthesis [28].
It is believed that FFAR2 is crucial in the human nutritional sensing system. Although studies on these receptors have suggested that they could be used to treat diabetes and other metabolic disorders [29], their utility in the treatment of CAD is still up for debate. More prospective studies are needed to look at its level among different forms of CAD, both in the disease's early and late phases to see if elevated expression levels could be a target for early therapy in patients with SA and to prevent cardiac complications in patients with UA.
Conclusion
The TL1A gene was upregulated and the FFAR2 gene was downregulated in the circulating blood of SA and UA patients. TL1A and FFAR2 are sensitive and specific biomarkers for discriminating SA from controls and for distinguishing UA from SA. TL1A and FFAR2 are independent risk factors for SA. However, TL1A and not FFAR2 is an independent predictor for UA. Our findings suggest that TL1A and FFAR2 are hopeful and useful blood biomarkers for the early detection of SA and UA, as well as for the early management and prevention of cardiac injury.
Availability of data and materials
Data available when required.
Abbreviations
- ACS:
-
Acute coronary syndrome
- AMI:
-
Acute myocardial infarction
- CAD:
-
Coronary artery disease
- DR3:
-
Death receptor 3
- FBG:
-
Fasting blood glucose
- FFAR2:
-
Free fatty acid receptor 2
- HDL-C:
-
High-density lipoprotein cholesterol
- hs-CRP:
-
High-sensitivity C-reactive protein
- IL-1:
-
Interleukin-1
- LDL-C:
-
Low-density lipoprotein cholesterol
- SA:
-
Stable angina
- TC:
-
Total cholesterol
- TG:
-
Triglycerides
- TL1A:
-
Tumor necrosis factor-like cytokine 1A
- TNF:
-
Tumor necrosis factor
- UA:
-
Unstable angina
References
Gorabi A, Sathyapalan T, Al-Rasadi K, Jamialahmadi T, Sahebkar A (2020) The role of MicroRNAs in regulating cytokines and growth factors in coronary artery disease: the ins and outs. J Immunol Res 2020:5193036
Ali Sheikh MS (2020) Diagnostic role of plasma MicroRNA-21 in stable and unstable angina patients and association with aging. Cardiol Res Pract 2020:9093151
Ji Q, Lin Y, Liang Z, Yu K, Liu Y, Fang Z et al (2014) Chemerin is a novel biomarker of acute coronary syndrome but not of stable angina pectoris. Cardiovasc Diabetol 13:145
Rahimzadeh M, Farshidi N, Naderi N, Farshidi H, Montazerghaem H (2020) Clinical significance of serum concentrations of neuregulin-4, in acute coronary syndrome. Sci Rep 10(1):5797
Chen X, Guo Y, Lai L, Zhang S, Li Z (2020) Intracoronary and peripheral blood levels of TNF-like Cytokine 1A (TL1A) in patients with acute coronary syndrome. Medicine 99(22):e20305
Akyüz A, ÖzkaramanlıGür D, Alpsoy Ş, Güzel S (2019) Tumour necrosis factor like cytokine 1A levels and lesion complexity in non-smoking patients with coronary artery disease. Biomark Biochem Indic Expos Response Suscep Chem 24(8):764–770
Ruan J, Meng H, Wang X, Chen W, Tian X, Meng F (2020) Low expression of FFAR2 in peripheral white blood cells may be a genetic marker for early diagnosis of acute myocardial infarction. Cardiol Res Pract 2020:3108124
Fihn SD, Gardin JM, Abrams J, Berra K, Blankenship JC, Dallas AP et al (2012) 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS guideline for the diagnosis and management of patients with stable ischemic heart disease: a report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines, and the American College of Physicians, American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Circulation 126(25):e354-471
Braunwald E, Antman EM, Beasley JW, Califf RM, Cheitlin MD, Hochman JS et al (2002) ACC/AHA 2002 guideline update for the management of patients with unstable angina and non-ST-segment elevation myocardial infarction–summary article: a report of the American College of Cardiology/American Heart Association task force on practice guidelines (Committee on the Management of Patients With Unstable Angina). J Am Coll Cardiol 40(7):1366–1374
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 25(4):402–408
Li X, Yang Y, Hao Y, Yang Y, Zhao J, Dou K (2013) Effects of pre-procedural statin therapy on myocardial no-reflow following percutaneous coronary intervention: a meta analysis. Chin Med J 126(9):1755–1760
Robinson S, Follo M, Haenel D, Mauler M, Stallmann D, Tewari M et al (2018) Droplet digital PCR as a novel detection method for quantifying microRNAs in acute myocardial infarction. Int J Cardiol 257:247–254
Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B et al (2002) TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity 16(3):479–492
Kim SH, Lee WH, Kwon BS, Oh GT, Choi YH, Park JE (2001) Tumor necrosis factor receptor superfamily 12 may destabilize atherosclerotic plaques by inducing matrix metalloproteinases. Jpn Circ J 65(2):136–138
Li XY, Hou HT, Chen HX, Wang ZQ, He GW (2018) Increased circulating levels of tumor necrosis factor-like cytokine 1A and decoy receptor 3 correlate with SYNTAX score in patients undergoing coronary surgery. J Int Med Res 46(12):5167–5175
Stamatelopoulos K, Georgiou S, Kanakakis I, Papamichael C, Oikonomidis N, Mantzou A et al (2015) Circulating levels of TNF-like cytokine 1A correlate with reflected waves and atherosclerosis extent and may predict cardiac death in patients with stable coronary artery disease. Cytokine 72(1):102–104
Liu C, Li XX, Gao W, Liu W, Liu DS (2014) Progranulin-derived Atsttrin directly binds to TNFRSF25 (DR3) and inhibits TNF-like ligand 1A (TL1A) activity. PLoS ONE 9(3):e92743
Richards LB, Li M, van Esch BC, Garssen J, Folkerts G (2016) The effects of short-chain fatty acids on the cardiovascular system. PharmaNutrition 4(2):68–111
Ma L, Wang T, Shi M, Fu P, Pei H, Ye H (2016) Synthesis, activity, and docking study of novel phenylthiazole-carboxamido acid derivatives as FFA2 agonists. Chem Biol Drug Des 88(1):26–37
Secor JD, Fligor SC, Tsikis ST, Yu LJ, Puder M (2021) Free fatty acid receptors as mediators and therapeutic targets in liver disease. Front Physiol 12:656441
Priyadarshini M, Navarro G, Layden BT (2018) Gut microbiota: FFAR reaching effects on islets. Endocrinology 159(6):2495–2505
Mishra SP, Karunakar P, Taraphder S, Yadav H (2020) Free fatty acid receptors 2 and 3 as microbial metabolite sensors to shape host health: pharmacophysiological view. Biomedicines. 8(6):154
Kimura I, Ichimura A, Ohue-Kitano R, Igarashi M (2020) Free fatty acid receptors in health and disease. Physiol Rev 100(1):171–210
Lymperopoulos A, Suster MS, Borges JI (2022) Short-chain fatty acid receptors and cardiovascular function. Int J Mol Sci 23(6):3303
Ang Z, Er JZ, Tan NS, Lu J, Liou YC, Grosse J et al (2016) Human and mouse monocytes display distinct signalling and cytokine profiles upon stimulation with FFAR2/FFAR3 short-chain fatty acid receptor agonists. Sci Rep 6:34145
Christiansen CB, Gabe MBN, Svendsen B, Dragsted LO, Rosenkilde MM, Holst JJ (2018) The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am J Physiol Gastrointest Liver Physiol 315(1):G53-g65
Batterham RL, Cowley MA, Small CJ, Herzog H, Cohen MA, Dakin CL et al (2002) Gut hormone PYY(3–36) physiologically inhibits food intake. Nature 418(6898):650–654
Nogal A, Valdes AM, Menni C (2021) The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 13(1):1–24
Fuller M, Li X, Fisch R, Bughara M, Wicksteed B, Kovatcheva-Datchary P et al (2016) FFA2 contribution to gestational glucose tolerance is not disrupted by antibiotics. PLoS ONE 11(12):e0167837
Acknowledgements
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AAK prepared the idea, designed the study, and did the statistical analysis; ST selected the patients and the control group; AAM prepared the idea, performed all the laboratory investigations, and interpreted patients’ data regarding each studied group. All authors wrote, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. The study was approved by the Bioethics Committee of Assiut University's Faculty of Medicine (IRB. no17300768). Informed consent was obtained from all individual participants included in the study.
Competing interests
There are no conflicts of interest declared by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kamel, A.A., Taha, S. & Mosa, A.A. Circulating expression patterns of TL1A and FFAR2 in patients with stable and unstable angina. Egypt J Med Hum Genet 24, 4 (2023). https://doi.org/10.1186/s43042-023-00386-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-023-00386-1