Abstract
The entomopathogenic genus Ophiocordyceps includes a highly diverse group of fungal species, predominantly parasitizing insects in the orders Coleoptera, Hemiptera, Hymenoptera and Lepidoptera. However, other insect orders are also parasitized by these fungi, for example the Blattodea (termites and cockroaches). Despite their ubiquity in nearly all environments insects occur, blattodeans are rarely found infected by filamentous fungi and thus, their ecology and evolutionary history remain obscure. In this study, we propose a new species of Ophiocordyceps infecting the social cockroaches Salganea esakii and S. taiwanensis, based on 16 years of collections and field observations in Japan, especially in the Ryukyu Archipelago. We found a high degree of genetic similarity between specimens from different islands, infecting these two Salganea species and that this relationship is ancient, likely not originating from a recent host jump. Furthermore, we found that Ophiocordyceps lineages infecting cockroaches evolved around the same time, at least twice, one from beetles and the other from termites. We have also investigated the evolutionary relationships between Ophiocordyceps and termites and present the phylogenetic placement of O. cf. blattae. Our analyses also show that O. sinensis could have originated from an ancestor infecting termite, instead of beetle larvae as previously proposed.
Similar content being viewed by others
INTRODUCTION
The genus Ophiocordyceps (Hypocreales, Ophiocordycipitaceae) comprises species typically pathogenic to insect hosts. Recently, however, there are reports of beneficial, endosymbiotic species of sap-sucking hemipterans hosts (Quandt et al. 2014; Gomez-Polo et al. 2017; Matsuura et al. 2018). The genus was erected by Petch (1931) to accommodate species of Cordyceps exhibiting clavate asci containing spores that do not disarticulate into partspores, contrasting with “the majority of the species of Cordyceps which have been described” at that time, exhibiting cylindrical asci and spores that readily disarticulate into numerous short partspores upon maturity. The diversity of Ophiocordyceps has been increasingly unraveled in the last decade, especially with discoveries of species associated with Hymenoptera, Lepidoptera and Hemiptera (Araújo et al. 2016, 2018; Luangsa-ard et al. 2018). However, our knowledge about species associated with blattodean insects (cockroaches and termites) is still restricted, especially regarding cockroach parasites. Currently, we know of only 11 species infecting termites, i.e. O. bispora (Stifler 1941); O. octospora (Blackwell and Gilbertson 1981); O. communis (Sung et al. 2007); O. asiatica, O. brunneirubra, O. khokpasiensis, O. koningsbergeri, O. mosingtoensis, O. pseudocommunis, O. pseudorhizoidea, and O. termiticola (Tasanathai et al. 2019). Furthermore, there are only three species described infecting cockroaches, i.e. O. blattae from Sri Lanka (formerly Ceylon) – (Petch 1931); O. blattarioides from Colombia – (Sanjuan et al. 2015) and O. salganeicola sp. nov. from Japan – this study).
The cockroaches (Blaberidae, Blattodea) are ubiquitous organisms occupying almost all habitats where insects occur (Schall et al. 1984). They play a key ecological role as decomposers, with many examples of species living within and feeding on rotting wood, including a lineage of social species that evolved into the termites (Bell et al. 2007; Maekawa et al. 2008; Bourguignon et al. 2018; Evangelista et al. 2019). Parasitism by microsporidians and protozoans on these insects is relatively common and known for more than a century (Crawley 1905; Woolever 1966; Purrini et al. 1988). However, the cases of infection by filamentous fungi on cockroaches have been rarely reported.
Although examples of Ophiocordyceps parasitizing Blattodea are scarce, the type of the genus is O. blattae infecting a cockroach identified as Blatta germanica (Fig. 1, adapted from Petch 1924). Only two specimens were originally collected in Sri Lanka (See Fig. 1a-d) with recent few records from Thailand (Luangsa-ard et al. 2018). Another example of an Ophiocordyceps infecting a cockroach is O. blattarioides (syn. Paraisaria blattarioides; Mongkolsamrit et al. 2019). This species was described from Colombia with records in Belize and tropical lowlands in the eastern (Amazonian) Ecuador (Sanjuan et al. 2015). Both O. blattae and O. blattarioides exhibit striking morphological and ecological differences. For example, O. blattae forms a cylindric ascoma producing elongated-fusoid ascospores, which do not disarticulate into part-spores, measuring 50–80 × 3–4 μm, while O. blattarioides has a stalk bearing a globoid fertile part at the tip, producing spores that disarticulate into part-spores of 6–12 × 1.5 μm. The host death location is also distinct with O. blattae occuring on the underside of leaves while O. blattarioides is found buried in the leaf litter. Besides these studies, there is no detailed information on the evolution and ecology of cockroach-associated entomoparasitic fungi.
Ophiocordyceps blattae and O. cf. blattae. a the original illustration of O. blattae showing the ascoma emerging laterally from the host thorax (Petch 1924). b and c (K98612 – holotype deposited in the Kew Gardens Fungarium). d and e Additional specimen collected by Petch in 1914, same location as the holotype K(M)264510. f O. cf. blattae from Thailand used in this study with two ascomata arising laterally on both sides (deposited at Biotech Fungal Collections as MY34765) (del. M. G. Moriguchi). a–e; images by Lee Davis, Royal Botanic Garden, Kew
In this study, we propose a new species of Ophiocordyceps that parasitizes two social wood-feeding cockroach species distributed in the southwestern part and Nansei Islands of Japan, both living inside decaying logs. We provide morphological, molecular and ecological data to support the new species proposal with insights into the evolutionary origins of the closely related parasitic fungi of cockroaches and termites. We also present, for the first time, the phylogenetic position of O. cf. blattae.
MATERIALS AND METHODS
Sampling
Surveys were undertaken in the Japanese warm temperate and subtropical evergreen forests mainly consisting of trees belonging to Fagaceae, Lauraceae and Theaceae in Kunigami-son, Okinawa, Yakushima, Kagoshima and Nobeoka, Miyazaki. The parasitized cockroach samples of two host species, namely Salganea esakii and S. taiwanensis, were mainly collected in the small humid valley or riparian forests where Castanopsis sieboldii, Distylium racemosum and Schefflera heptaphylla grow, but also in the secondary forest harboring Alnus japonica after the deforestation of Castanopsis in Okinawa. The specimens used in this study were always found hidden inside soft rotten logs or large fallen branches of those trees, with only the fungus emerging (Fig. 2). The infected cockroaches, and the substrata they were attached to, were collected in plastic containers and transported to the laboratory. Some specimens were investigated immediately after the collection, while others were dried and preserved for many years before being analyzed. The specimens were photographed individually, using a Canon 7D camera, equipped with an EF-100 mm macro lens or a MP-E 65 mm (5X) lens with a MT-24EX Canon macro lite flash attached.
Ophiocordyceps salganeicola emerging from Salganea esakii in its natural habitat in Yakushima island, Kagoshima. a and b Visible part of the ascoma, emerging from a hole in the wood. c and d Hosts buried in the wood, visible only after digging a few centimeters. e Infected host in its very rare nymphal stage
Collections
We collected 26 samples in two locations in Okinawa, 27 in Yakushima island, Kagoshima, and five in Miyazaki. Four specimens from Yonahadake and Kunigami (Okinawa) and two from Kagoshima were used for DNA extraction and sequencing for each prefecture (see Fig. 3, Table 1). Almost all fungal specimens were collected from adults of S. esakii and S. taiwanensis between April – June from 2004 to 2019.
Southwestern part of Japan showing collection sites of O. salganeicola specimens used in this study. Specimen details are indicated in Table 1. The map was retrieved and edited from Geospatial Information Authority of Japan
Morphological studies
For macro-morphological characterization, specimens were examined using a stereoscopic microscope (Leica S8 APO) and sorted for further micro-morphological investigation. The characters investigated were ascomatal size, color, position, presence/absence and characterization of asexual morphs and perithecial insertion (e.g. immersed, semi-immersed, erumpent, superficial). For micro-morphological characterization, either free-hand or cryo-sectioning of the ascoma was performed using a Leica CM1850 Cryostat. Samples were mounted on a slide with plain lactic acid or lacto-fuchsin (0.1 g of acid fuchsin in 100 mL of lactic acid) for light microscopy examination using a Nikon Eclipse Ni-U. A minimum of 50 ascospores were measured for morphological comparison. The illustrations of fungal specimens were drawn based on the observation of photographs using drawing pens 0.13 mm and 0.2 mm (Rotring, Hamburg, Germany), painted by watercolors (HOLBEIN Art Materials Inc., Osaka, Japan) and scanned for imaging (Fig. 4). We also present the morphological comparison between Ophiocordyceps species infecting cockroaches and termites (Table 2).
Ophiocordyceps salganeicola at various stages of development (del. M. G. Moriguchi). a Sexual morph of O. salganeicola growing on Salganea esakii collected in Miyazaki, 22 Jun. 2005. b Sexual morph on S. esakii collected in Yakushima, 17 Jun. 2017. with a close-up illustration of perithecia. c Early-stage specimen. d Mid-stage specimen (Mori04; see Tables 1 and 2) collected in Katsuu-dake, Nago-shi, Okinawa, 23 Apr. 2016. e Mature specimen from Kunigami-son, Okinawa, collected 29 May 2016
DNA extraction, PCR and sequencing
All specimens used in this study were collected in their natural habitat. The material was preserved either dried or in ethanol and DNA extractions were performed with the following protocol: Parts of fungal tissues were removed from the host, placed in 1.5 ml Eppendorf tubes with 100–200 μl of CTAB readily after its collection and stored at room temperature, or entire samples were immersed in 70% ethanol and stored in the freezer. For DNA extraction, the samples were ground mechanically with 400 μl of CTAB and incubated at 60 °C for 20 min and centrifuged for 10 min at 14,000 rpm. The supernatant (approx. 400 μl) was transferred to a new 1.5 ml Eppendorf tube, mixed with 500 μl of 24:1 chloroform: isoamyl-alcohol (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) and mixed by inverting. The mix was then centrifuged for 20 min at 14,000 rpm and the supernatant transferred to a new 1.5 ml Eppendorf tube and further cleaned using the GeneCleanIII kit (MP Biomedicals, Santa Ana, CA, USA), following the recommended protocol. The only step modified was the addition of 30 μl of GlassMilk per sample, instead of the recommended 10 μl, aiming to increase yield.
Five loci were used in the analyses, i.e. small subunit nuclear ribosomal DNA (SSU), large subunit nuclear ribosomal DNA (LSU), translation elongation factor 1-α (TEF) and the largest and second largest subunits of RNA polymerase II (RPB1 and RPB2 respectively) with a total read length of 4815 bp. The primers used were, SSU: 82F (5′-GAAACTGCGAATGGCT-3′) and 1067R (5′-TMTCGTAAGGTGCCGA-3′) (Matsuura et al. 2018); LSU: LR0R (5′-ACCCGCTGAACTTAAGC-3′) and LR5 (5′-TCCTGAGGGAAACTTCG-3′) (Vilgalys and Sun 1994); TEF: 983F (5′-GCYCCYGGHCAYCGTGAYTTYAT-3′) and 2218R (5′- ATGACACCRACRGCRACRGTYTG-3′); cRPB1: (5′-CCWGGYTTYATCAAGAARGT-3′) and RPB1Cr (5′-CCNGCDATNTCRTTRTCCATRTA-3′) (Castlebury et al. 2004). RPB2: fRPB2-5F:(5′-GAYGAYMGWGATCAYTTYGG-3′) and fRPB2-7cR (5′- CCCATRGCTTGTYYRCCCAT − 3′) (Liu et al. 1999).
Each 20 μl-PCR reaction contained 10 μl of Ampdirect® Plus 2x (Shimazu Corp., Kyoto, Japan), 0.6 μl of each forward and reverse primers (10 mM), 1 μl of DNA template, 0.1 TaKaRa Ex Taq DNA Polymerase (Takara Bio Inc., Kusatsu, Shiga, Japan) and 8.7 μl of Ultra Pure Distilled Water (Thermo Fisher Scientific Inc., Waltham, MA, USA). The PCR reactions were placed in an Astec PC-818 thermocycler under the following conditions: for SSU and LSU (1) 2 min at 95 °C, (2) 10 cycles of denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s, and extension at 72 °C for 2 min, followed by (3) 25 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 2 min and (4) 3 min at 72 °C. For TEF and RPB1(1) 2 min at 95 °C, (2) 10 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 40 s, and extension at 72 °C for 1 min 30 s, followed by (3) 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 40 s, and extension at 72 °C for 1 min 30 s and (4) 3 min at 72 °C. Each PCR reaction was partially electrophoresed and the rest was cleaned by adding 3.0 μl of enzymatic PCR clean-up reagent, consisting of 0.1 μl of Exonuclease I (New England BioLabs, Ipswich, MA, USA) and 0.1 μl of alkaline phosphatase (shrimp) (Takara Bio Inc., Kusatsu, Shiga, Japan), incubated at 37 °C for 20 min and 80 °C for 15 min in the thermocycler. The processed PCR products were directly sequenced by a capillary DNA sequencer, Genetic Analyzer 3130xl (Thermo Fisher Scientific Inc., Waltham, MA, USA) at C-RAC of the University of the Ryukyus.
Phylogenetic analyses
The raw sequence reads (.ab1 files) were edited manually using Geneious 11.1.5 (https://www.geneious.com). Individual gene alignments were generated by MAFFT (Katoh and Standley 2013). The alignment of every gene was improved manually, annotated and concatenated into a single combined dataset using Geneious 11.1.5. Ambiguously aligned regions were excluded from phylogenetic analysis and gaps were treated as missing data. The final alignment length was 4629 bp: 1020 bp for SSU, 870 bp for LSU, 967 bp for TEF, 683 bp for RPB1 and 1089 for RPB2. Maximum likelihood (ML) analysis was performed with RAxML version 8.2.4 (Stamatakis 2014) on a concatenated dataset containing all five genes. The dataset consisted of 11 data partitions, 2 for SSU and LSU, and 9 for each codon position of the three protein coding genes: TEF, RPB1 and RPB2. The GTRGAMMA model of nucleotide substitution was employed during the generation of 1000 bootstrap replicates. The sequences for all Ophiocordyceps used in this study are presented in Table 3.
Ancestral character state reconstruction (ACSR)
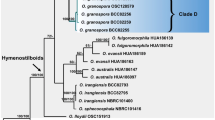
To understand the evolutionary pathways of host association of Ophiocordyceps and blattodean insects, we conducted ancestral character reconstruction in Mesquite (Maddison and Maddison 2018) of the whole genus, using the best-scoring ML tree produced in RAxML (Stamatakis 2014). We coded each taxon based on host association (8 categories: Acari, Coleoptera, Hymenoptera, Lepidoptera, Hemiptera, Orthoptera and Blattodea divided into cockroaches and termites – Fig. 5). We used maximum likelihood model MK1, as implemented in Mesquite 3.51 (Maddison and Maddison 2018). Only nodes presenting > 70% of probability were displayed and used to color-code the branches on the Fig. 5. Nodes below this limit were treated as ambiguous and displayed as dashed lines.
Maximum likelihood tree of Ophiocordycipitaceae obtained from RAxML analyses based on a concatenated set of 5 genes (SSU, LSU, TEF, RPB1 and RPB2). Colored branches reflect Ancestral Character State Reconstruction (ACSR) analyses based on host associations (See legend at the bottom-left) and pie-charts represent the probability for the association with host orders. Dashed lines indicate ambiguous association. Host pictures by Alex Wild and Shizuma Yanagisawa
TAXONOMY
Ophiocordyceps salganeicola Araújo, Moriguchi & Matsuura, sp. nov.
Ophiocordyceps salganeicola on Salganea taiwanensis (dried specimen) from Kunigami-son, Okinawa (TNS-F-60532). a Two ascomata arising from S. taiwanensis. b Close-up of dried ascoma. c Cross section of ascoma showing the perithecial arrangement. d Ascus with spirally twisted ascospores. e Perithecial ostiole. f and g 8-celled ascospore. Scale bars = c 200 μm, d 15 μm, e 10 μm, f and g 20 μm. Presented as JPMA124 in
Ophiocordyceps salganeicola on Salganea esakii (fresh specimen) from Yakushima, Kagoshima (TNS-F-91239). a Salganea esakii with a single robust ascoma; b Close-up showing early stage ascoma arising from ventral pronotum; c and d Close-up of ascoma; e Cross-section of ascoma; f and g Perithecia; h Ascospores within ascus; i and j 8-celled ascospores. Presented as JPMA106 in Fig. 5
Mycobank MB836091.
Etymology: Named after the host genus Salganea.
Diagnosis: Ophiocordyceps salganeicola can be easily differentiated from other closely-related species by its unique host association, ascomatal morphology and its strict distribution across the Southern Islands of Japan. Other closely related species are associated with termites, mites and hemipterans and exhibit completely different macro morphology, being easily distinguished still in the field.
Type: Japan: Okinawa: Kunigami-son, Yonahadake, 26°43′45.0″N 128°12′48.2″E, on Salganea taiwanensis (Blattodea, Blaberidae), 21 June 2017, M.G. Moriguchi (TNS-F-60532 – holotype).
Description: External mycelium sparse, light to dark brown, arising from the host’s sutures. Stromata one or two, 1–7 cm long, 1.3–5 mm thick, cream to light or dark brown, clavate to cylindrical in shape. Perithecia immersed, usually covering the apical part descending tol about the middle of the stromata, immersed to semi-immersed, ovoid to flask-shaped, (325–) 365 (− 408) × 100–140 μm. Asci hyaline, elongated clavate, 150–200 × 7–11 μm with prominent cap, 8-spored. Ascospores hyaline, 70–100 × 3 μm, 7-septate, not disarticulating into part-spores. Asexual morph: hirsutella-like phialides occurring sparsely on the surface of the stromata where perithecia are absent, 7–16 × 6–8 μm with neck measuring 18–30 × 5–7.5. Conidia: ovoid, 7 × 5 μm, hyaline to pale brown.
Host: Salganea taiwanesnsis and S. esakii.
Habitat: Forests of Miyazaki Prefecture, Yakushima and Okinawa islands of Japan. On remains of the hosts inside rotten logs, with only the fungus sporophores emerging.
Distribution: Only currently known from Japan.
Other specimen examined: Japan: Shirakawayama, Issou, Yakushima, Kagoshima 30°20′58.0″N 130°36′35.”E, on Salganea esakii (Blattodea, Blaberidae), 22 June 2019, N. Kinjo (TNS-F-91239 – paratype) (as JPMA106 see Fig. 7).
RESULTS
Molecular phylogeny and evolutionary origins of cockroach-associated Ophiocordyceps
We obtained 20 new sequences from five specimens of O. salganeicola (Fig. 3, Table 3). Our phylogenetic analysis is in accordance with previously published Ophiocordyceps topologies (Quandt et al. 2014; Sanjuan et al. 2015; Araújo et al. 2018; Tasanathai et al. 2019). All O. salganeicola specimens we collected, from different parts of Japan and infecting two species of Salganea, clustered together as a single species with a high degree of genetic similarity with a long branch (Fig. 5). It formed a monophyletic group with another cockroach-associated species, O. blattae, which is the type species for Ophiocordyceps. This is the first time O. cf. blattae is included in a phylogenetic study.
Our results indicate that Ophiocordyceps originated from a beetle-associated ancestor (72% ACSR), corroborating previous studies (Araújo and Hughes 2019). For the cockroach parasites, we found at least two independent origins within Ophiocordyceps, one within the Paraisaria clade, i.e. Paraisaria blattarioides (Fig. 5 node A), and the other within the hirsutelloid species, i.e. O. salganeicola and O. blattae (Fig. 5 node B). The ancestral host association for the cockroach-associated Paraisaria lineage was ambiguously recovered, while for the hirsutelloid cockroach-associated species our data show it has originated likely from a termite-associated ancestor, although this is not strongly supported (44% ACSR). We also found that the association with termites is older than cockroaches, evolving independently at least twice (Fig. 5 nodes C and D). The oldest, would have arisen from beetles to termites (65% ACSR, Fig. 5 node C). However, the origins of O. brunneirubra remains uncertain as part of the ancient termite-associated lineage (Fig. 5 node C) or if it jumped more recently from Hymenoptera to termites (Fig. 5 Node D).
The Paraisaria clade is an ecologically heterogeneous group composed of species parasitic on Coleoptera, Orthoptera, Lepidoptera, and Hemiptera (Mongkolsamrit et al. 2019). Our ACSR analysis provided weak resolution for the origins of P. amazonica/P. blattarioides/P. gracilis clade with 50.1% for Orthoptera, 25.9% for Blattodea (cockroaches) and 10.9% for Lepidoptera (Fig. 8 Node A). Our data also did not provide strong support for the ancestor of P. blattarioides with 51.1% for Blattodea (cockroaches), 21% for Orthoptera and 20.6% for Lepidoptera (Fig. 8 Node A). Nevertheless, the whole Paraisaria lineage was strongly supported as having evolved from a beetle parasite (Fig. 8 Node F, ACSR = 81.1%).
Conversely, for the novel clade composed of O. salganeicola and O. blattae, our results suggest (BS=87; ACSR=72.4%) that it evolved from an ancestral parasite on termites (Fig. 8 node E). Ophiocordyceps salganeicola/blattae was retrieved as a sister group to a clade composed mostly by termite (Blattodea, Termitidae) parasites with species associated with hemipterans (Pseudococcidae) and mites (Acari, Eriophyidae). According to our results, all host switches in this clade occurred from termites (i.e. termites to Coleoptera, termites to Hemiptera, termites to Acari and termites to cockroaches). Unexpectedly, our analyses also suggest (ACSR=61.7%) the clade composed by parasites of Coleoptera, Lepidoptera, and Hemiptera, including the economically and culturally important O. sinensis, could have originated from an ancestor infecting termites, instead of beetle larvae as previously proposed (Araújo and Hughes 2019).
DISCUSSION
Ecology and natural history of Salganea–Ophiocordyceps relationships
The insect cuticle represents a formidable barrier to infection by bacterial and fungal pathogens with relatively few having managed to cross it. Once inside the insect, innate immunity provides further challenges to an invading pathogen (Evans 1988). However, once these have been overcome the insect body provides a stable environment for their development. This is particularly true for colonial cockroaches that spend most of their lives protected inside nests, for example some wood-feeding species within the families Cryptocercidae and Blaberidae, specifically the subfamily Panesthiinae (Panesthiini, Ancaudeliini, Caepariini, and Salganeini). Among those groups, one of the most well-known social cockroaches is the genus Salganea, comprised of about 50 species (Beccaloni 2007; Bell et al. 2007; Wang et al. 2014). All the known species within the genus live within and feed on decaying wood, building chambers and galleries inside hardwood or coniferous logs that may take decades to degrade (Maekawa et al. 2008), providing long-term stable homeostatic conditions. Such a protected environment certainly benefits indirectly the fungal parasites that are already inside the host body. An exposed cadaver on the forest floor would be much more susceptible to being scavenged by animals or consumed by other microorganisms.
Salganea species form social groups, composed mostly of biparental families, consisting of a male-female and their offspring (Maekawa et al. 2008). Sociality endows insects with advantages such as increased efficiency of brood care, foraging and anti-predator defenses. However, infectious diseases can potentially spread more easily within a colony because of their high densities, frequent social contact and also because group members are often close relatives and thus susceptible to the same parasitic infections (Cremer et al. 2007). Therefore, it is surprising that only three species of Ophiocordyceps, a common and widespread genus of entomopathogenic fungi, have been recorded infecting the equally diverse and globally distributed cockroaches (Bourguignon et al. 2018). Ophiocordyceps species infecting social insects, notably ants, are one of the most broadly distributed and ubiquitous entomopathogenic fungi in tropical forests worldwide (Araújo et al. 2015, 2018). They often form epizootic events, in which hundreds of infected ants can be found in a small patch of forest (Evans and Samson 1982; Pontoppidan et al. 2009). On the other hand, however, Ophiocordyceps on social cockroaches are rare in Japan and only one or two infected individuals are collected in the same log, despite the ubiquity and abundance of hosts in one area.
Based on our extensive field surveys we found that ascomata of O. salganeicola in Okinawa start to emerge in decaying logs in early April. However, they seem to require at least a few months to become mature and develop the sexual morph in the field. This development occurs in parallel with the mating season of the host cockroaches from April to July when newly emerged adults leave their logs and parents, fly, mate and burrow into a new nest (Osaki Haruka, personal communication). Presumably, these young adults might become infected by the ascospores/conidia of O. salganeicola during colonization of a new log. The host is then later killed and consumed by the fungal parasite, eventually producing new fruiting bodies in the next mating season. On the other hand, there has been no record of an infected nymph in 26 fungal specimens observed in Okinawa, but only a single infected nymph (see Fig. 2e) out of more 27 specimens in Yakushima between 2015 and 2019, suggesting the outbreak of this fungus within an established colony is rather rare and the primary targets are likely newly emerged adults. However, this proposed life-cycle of O. salganeicola is only hypothetical and requires periodical field observations in the same ecological habitat along with host insect behaviors (Maekawa et al. 2008), in order to determine how and when the fungus infects and kills the host.
Does O. salganeicola manipulate host behavior before death?
The behavior manipulation caused by Ophiocordyceps fungi on their hosts is a striking phenomenon, especially in species associated with ants, the so-called “zombie-ant fungi” (Evans et al. 2011; Araújo et al. 2018). It has been posited that species within the Ophiocordyceps unilateralis core clade infecting Camponotini ants evolved such an ability as a response to the strong social immunity displayed by ant societies that prevents fungal transmission and development inside the colony (Araújo and Hughes 2019). Conversely, as far as we know, there is no evidence of social cockroaches recognizing the infected members of their colony, except for the parental and sibling’s grooming behavior that might fend off superficial parasites. Thus, fungal infection, development and transmission could potentially occur in the same log where other members of the colony still inhabit, in which no drastic behavior manipulation is needed in order to remove the host from its nest and thus complete the parasite’s life-cycle. However, there is a possibility of a subtle manipulation.
Salganea cockroaches burrow and nest deep inside the trunk, only becoming exposed to the external environment in the mating season, whereas the Ophiocordyceps-infected ones are found only a few centimeters below the wood surface (Fig. 2). Thus, we posit that the fungus might potentially be able to manipulate host’s behavior by leading to a migration towards a more superficial layer inside the log. This host migration could be stimulated by the need for a more oxygen- and water-rich stratum and/or attraction to the light coming from an opening, through which the fungal ascoma emerge and disperse its spores (Fig. 2a–b). Further studies are needed to test this hypothesis.
Host association, speciation, and distribution
While some distinct, mostly macro-, morphological features can be observed in O. salganeicola infecting both host species that diverged from a single ancestor around 4–5 mya (Maekawa et al. 1999; Maekawa and Matsumoto 2003), nucleotide sequences of fungal specimens are highly conserved among diverse strains in wide range of geographic regions and islands. The number of polymorphic sites within all aligned five gene sequences from multiple samples (Table 3) was only two out of 4202 and both were synonymous. Thus, geographic and reproductive isolation of the fungal strains may have not yet resulted in the allopatric speciation of O. salganeicola. The long branch length and high host specificity to the genus Salganea indicate a long co-evolutionary relationship with the host populations and hence unlikelihood of recent host jumping (Fig. 5 node B). However, we still do not know whether the single parasite strain can only persist in one geographic region and/or island infecting the same host populations over generations, or jump across multiple closely related host populations and species horizontally even after such a long geographic isolation of the Ryukyu Islands. In some of these islands such as Amamioshima and Tokunoshima in Kagoshima and Ishigaki-jima and Iriomote-jima in Okinawa, there has been no collection record of O. salganeicola, suggesting their current absence. These questions on the evolution of host associations of O. salganeicola in the Japanese archipelago of Ryukyus deserve particular attention for studying host-parasite co-evolution in the context of island biology, which possibly can be tested by artificial infection experiments using the field-collected fungal ascoma and laboratory-reared Salganea colonies from different islands. Furthermore, S. taiwaensis and the other Salganea cockroach species are distributed not only in Japan, but also in wide greographic regions in South, Southeast and East Asia (Wang et al. 2014). Additional screening of Salganea and related host species for entomopathogens in Asia-Pacific regions might unravel the phylogeography and evolutionary origins of cockroach-Ophiocordyceps associations.
Ophiocordyceps cf. blattae
Ophiocordyceps blattae was described by Petch (1924) and was collected at Hakgala (Sri Lanka, formerly Ceylon) only twice (Fig. 1). A rare species, it was originally collected on cockroaches attached by fungal structures to the underside of leaves, exhibiting a cylindric, grey to lavender ascoma. The specimen we used in this study exhibited many similarities with the type specimen. For example, it also infects a very similar species of cockroach, kills its hosts on the underside of leaves, attached by fungal structures, and the ascomata emerge laterally from the host’s thorax. Furthermore, the ascomata are very similar in macro-morphological features. Unfortunately, we could not assess the micro-morphological features in this study and thus, to avoid any ambiguities, we are calling our material O. cf. blattae. We made efforts to sequence the original material from 1924, but all our attempts failed. As this is the type species of the genus Ophiocordyceps, further efforts are needed to fix the phylogenetic placement of the type species and thus, re-discuss the systematics of the whole genus. We have considered to propose the specimen used in this study as the epitype of O. blattae, however, since it was collected in Thailand, not Sri Lanka as it was originally found by Petch, future efforts might address this issue. However, herein we provide a good perspective for future efforts and revealed another clade likely bearing cryptic species within Ophiocordyceps on cockroaches.
CONCLUSION
Japan may harbour one of the richest reservoirs of entomopathogenic fungi in the world. In no other country is there an amateur society devoted to collecting and illustrating them. We still know very little about these organisms and their ecological roles in the environment and dynamic associations with host insects, including blattodean-associated ones such as O. salganeicola. Therefore, it is crucial to understand their true diversity with the invaluable help by such amateur and professional mycologists. As we move forward and describe more and more species through microscopic and molecular tools, new insights into the evolutionary origins of these organisms are being revealed, as well as their ecological associations with the insect hosts. Currently, there is still a substantial gap in our knowledge about insect ecology and fungal biology within the context of host-parasite interactions and their life-cycles. In this study, our goal was to describe an ecologically rare fungal species parasitizing unique social insects, and to provide some insights into their evolution by considering natural histories of both the parasite and its host. Thereby, our study contributes to the understanding of one of the most prolific and diverse groups of entomopathogenic fungi, the genus Ophiocordyceps, and incidentally shed new light on the origins of the economically important O. sinensis.
Availability of data and materials
The dataset analyzed in this study is available from the corresponding author upon request.
Abbreviations
- CTAB:
-
Cetyl Trimethyl Ammonium Bromide
- Min:
-
Minute
- Sec:
-
Seconds
- ml:
-
Milliliter
- mm:
-
Millimeter
- RAxML:
-
Randomized Axelerated Maximum Likelihood
- rpm:
-
Rotations per minute
- μl:
-
Microliter
References
Araújo JPM, Evans HC, Geiser DM, Mackay WP, Hughes DP (2015) Unravelling the diversity behind the Ophiocordyceps unilateralis (Ophiocordycipitaceae) complex: three new species of zombie-ant fungi from the Brazilian Amazon. Phytotaxa 220:224–238
Araújo JPM, Evans HC, Kepler R, Hughes DP (2018) Zombie- ant fungi across continents: 15 new species and new combinations within Ophiocordyceps. I. Myrmecophilous hirsutelloid species. Studies in Mycology 90:119–160
Araújo JPM, Hughes DP (2016) Diversity of entomopathogenic fungi: which groups conquered the insect body? Advances in Genetics 94:1–39
Araújo JPM, Hughes DP (2019) Zombie-ant fungi emerged from non-manipulating, beetle-infecting ancestors. Current Biology 29:3735–3738
Beccaloni GW (2007) Blattodea species file online. Version 5.0/5.0. World Wide Web electronic publication http://Blattodea.SpeciesFile.org. Accessed 8 July 2020 (A regularly updated world catalogue of extant Blattodea excluding the Termitoidae termites)
Bell WJ, Roth LM, Nalepa CA (2007) Cockroaches: ecology, behavior, and natural history. The Johns Hopkins University Press, USA
Blackwell M, Gilbertson RL (1981) Cordycepioideus octosporus, a termite suspected pathogen from Jalisco, Mexico. Mycologia 73:358–362
Bourguignon T, Tang Q, Ho SYW, Juna F, Wang Z, Arab DA, Cameron SL, Walker J, Rentz D, Evans TA, Lo N (2018) Transoceanic dispersal and plate tectonics shaped global cockroach distributions: evidence from mitochondrial phylogenomics. Molecular Biology and Evolution 35:970–983
Castlebury LA, Rossman AY, Sung G-H, Hyten A, Spatafora JW (2004) Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycological Research 108:864–872
Conlon BH, Mitchell J, de Beer ZW, Carøe C, Gilbert MT, Eilenberg J, Poulsen M, de Fine Licht HH (2017) Draft genome of the fungus-growing termite pathogenic fungus Ophiocordyceps bispora (Ophiocordycipitaceae, Hypocreales, Ascomycota). Data Brief. 11:537–542
Crawley H (1905) Coelosporidium blattellae, a new sporozoan parasite of Blattella germanica. Academy of Natural Sciences of Philadelphia 57:158–161
Cremer S, Armitage SAO, Schmid-Hempel P (2007) Social immunity. Current Biology 17:693–702
Evangelista DA, Wipfler B, Béthoux O, Donath A, Fujita M, Kohli MK, Legendre F, Liu S, Machida R, Misof B, Peters RS, Podsiadlowski L, Rust J, Schuette K, Tollenaar W, Ware JL, Wappler T, Zhou X, Meusemann K, Simon S (2019) An integrative phylogenomic approach illuminates the evolutionary history of cockroaches and termites (Blattodea). Proceedings of the Royal Society B 286:1–9
Evans HC (1988) Coevolution of entomogenous fungi and their insect hosts. In: Pirozynski KA, Hawksworth DL (eds) Coevolution of fungi with plants and animals. London Academic Press, London, pp 149–111
Evans HC, Elliot SL, Hughes DP (2011) Hidden diversity behind the zombie-ant fungus Ophiocordyceps unilateralis: four new species described from carpenter ants in Minas Gerias, Brazil. PLoS One 6:e1024
Evans HC, Samson RA (1982) Cordyceps species and their anamorphs pathogenic on ants (Formicidae) in tropical forest ecosystems. I. The Cephalotes (Myrmicinae) complex. Transactions of the British Mycological Society 79:431–453
Gomez-Polo P, Ballinger MJ, Lalzar M, Malik A, Ben-Dov Y, Mozes-Daube N, Perlman SJ, Iasur-Kruh L, Chiel E (2017) An exceptional family: Ophiocordyceps-allied fungus dominates the microbiome of soft scale insects (Hemiptera: Sternorrhyncha: Coccidae). Molecular Ecology 26:5855–5868
Katoh K, Standley KDM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30:772–780
Kepler RM, Kaitsu Y, Tanaka E, Shimano S, Spatafora JW (2011) Ophiocordyceps pulvinata sp. nov., a pathogen of ants with a reduced stroma. Mycoscience 52:39–47
Liu YL, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase IIsubunit. Molecular Biology and Evolution 16:1799–1808
Luangsa-ard JJ, Tasanathai K, Thanakitpipattana D, Khonsanit A, Stadler M (2018) Novel and interesting Ophiocordyceps spp. (Ophiocordycipitaceae, Hypocreales) with superficial perithecia from Thailand. Studies in Mycology 89:125–142
Maddison WP, Maddison DR (2018) Mesquite: a modular system for evolutionary analyses, version 3.51. http://www.mesquitweproject.org
Maekawa K, Lo N, Kitade O, Miura T, Matsumoto T (1999) Molecular phylogeny and geographic distribution of wood-feeding cockroaches in east Asian Islands. Molecular Phylogenetic and Evolution 13:360–376
Maekawa K, Matsumoto T (2003) Biogeography of Japanese wood-feeding cockroaches genus Salganea and Panesthia (Blaberidae: Panesthiinae). Molecular Phylogenetics and Evolution 27:156–159
Maekawa K, Matsumoto T, Nalepa CA (2008) Social biology of the wood-feeding cockroach genus Salganea (Dictyoptera, Blaberidae, Panesthiinae): did ovoviviparity prevent the evolution of eusociality in the lineage? Insectes Sociaux 55:107–114
Matsuura Y, Moriyama M, Łukasik P, Vanderpool D, Tanahashi M, Meng XY, McCutcheon JP, Fukatsu T (2018) Recurrent symbiont recruitment from fungal parasites in cicadas. Proceedings of National Academy of Sciences USA 115:E5970–E5979
Mongkolsamrit S, Noisripoom W, Arnamnart N, Lamlertthon S, Himaman W, Jangsantear P, Samson RA, Luangsa-ard JJ (2019) Resurrection of Paraisaria in the Ophiocordycipitaceae with three new species from Thailand. Mycological Progress 18:1213–1230
Ochiel GS, Evans HC, Eilenberg J (1997) Cordycepioideus, a pathogen of termites in Kenya. Mycologist 11:7–9
Petch T (1924) Studies in entomogenous fungus. IV. Some Ceylon Cordyceps. Transactions of the British Mycological Society 10:28–45
Petch T (1931) Notes on entomogenous fungi. Transactions of the British Mycological Society 16:55–75
Pontoppidan M-B, Himaman W, Hywel-Jones NL, Boomsma JJ, Hughes DP (2009) Graveyards on the move: the spatio-temporal distribution of dead Ophiocordyceps-infected ants. PLoS One 4(3):e4835
Purrini K, Weiser J, Kohring GW (1988) Coelosporidium tangae n. sp. (Protista), a new protist parasitizing a natural population of a field cockroach, Blatta sp. (Blattaria). Archiv für Protistenkunde 136:273–281
Quandt CA, Kepler RM, Gams W, Araújo JPM, Ban S, Evans HC, Hughes D, Humber R, Hywel-Jones N, Li Z, Luangsa-ard JJ, Rehner SA, Sanjuán T, Sato H, Shrestha B, Sung G-H, Yao Y-J, Zare R, Spatafora JW (2014) Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus 5:121–134
Sanjuan TI, Fanco-Molano AE, Kepler RM, Spatafora JW, Tabima J, Vasco-Palacios AM, Restrepo S (2015) Five new species of entomopathogenic fungi from the Amazon and evolution of neotropical Ophiocordyceps. Fungal Biology 119:901–916
Schall C, Gautier JY, Bell WJ (1984) Behavioural ecology of cockroaches. Biological Reviews 59:209–254
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stifler CB (1941) A new genus of Hypocreales. Mycologia 33:82–86
Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57:5–59
Tasanathai K, Noisripoom W, Chaitika T, Khonsanit A, Hasin S, Luangsa-ard JJ (2019) Phylogenetic and morphological classification of Ophiocordyceps species on termites from Thailand. MycoKeys 56:101–129
Vilgalys R, Sun BL (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proceedings of National Academy of Sciences USA 91:4599–4603
Wang X, Shi Y, Wang Z, Che Y (2014) Revision of the genus Salganea Stål (Blattodea, Blaberidae, Panesthiinae) from China, with descriptions of three new species. ZooKeys 412:59–87
Woolever P (1966) Life history and electron microscopy of a haplosporidian, Nephridiophaga blattellae (Crawley) n. comb., in the Malpighian tubules of the German cockroach, Blattella germanica (L.). Journal of Protozoology 13:622–642
Acknowledgments
We thank Shuichi Kurogi and Hiroaki Yamashita for sample collection in Miyazaki and Kagoshima, Shizuma Yanagisawa for Salganea cockroach photos, Haruka Osaki for the information on the host insect behaviors, Gaku Tokuda, Kiyoto Maekawa, and Christine Nalepa for the advice on the insect taxonomy and ecology, Pepijn Kooij and Lee Davis from Kew Gardens for the efforts to get DNA data from the holotype of O. blattae and Jennifer Luangsa-ard and Wasana Noisripoom for providing DNA sequences of O. cf. blattae found in Thailand. We would like to thank the reviewers for the very helpful suggestions to this manuscript. The authors also would like to highlight the extreme importance of work done previously in Japan, especially by Daisuke Shimizu and Yosio Kobayasi, who have inspired several generations of professional and amateur mycologists with their art and science.
Funding
J.P.M.A. conducted research as an invited foreign researcher at the University of the Ryukyus. This study was supported by JSPS Grant-in-Aid KAKENHI Grant No. 18 K19217, 18KK0211 and by a general research grant of the Institute of Fermentation, Osaka (IFO) to Y.M.
Author information
Authors and Affiliations
Contributions
J.P.M.A. conducted microscopy and molecular work, analyzed the data, photography, created and designed the figures and wrote the manuscript. M.G.M. collected samples, made the illustrations, provided important field observations and contributed with the discussion. S.U. collected samples and provided some microscopic images. N.K. collected samples and provided in situ images. Y.M. observed the natural habitats, conducted molecular work, analyzed the data and wrote the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Araújo, J.P.M., Moriguchi, M.G., Uchiyama, S. et al. Ophiocordyceps salganeicola, a parasite of social cockroaches in Japan and insights into the evolution of other closely-related Blattodea-associated lineages. IMA Fungus 12, 3 (2021). https://doi.org/10.1186/s43008-020-00053-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43008-020-00053-9