Abstract
Background
Hospital sewage is a significant reservoir of antimicrobial-resistant pathogens and genes that pose a huge public health threat. In this study, we determined the occurrence of multidrug-resistant Escherichia coli and Klebsiella pneumoniae in sewage flowing from a referral hospital through the urban sewage system to the point of discharge in the Indian Ocean.
Results
A total of 400 sewage samples were collected, yielding 517 isolates. Of these, 32.3% (167/517) were from hospital sewage, while 67.7% (350/517) were from the community. E. coli was the most common isolate (44.5% (230/517)), followed by K. pneumoniae at 27.3% (141/517), and other gram-negative bacteria constituted 28.2% (146/517) of the isolates. Multidrug resistance (MDR) was seen in 80.9% (186/230) E. coli and 71.6% (101/141) K. pneumoniae. Of the MDR isolates, 27.2% (78/287) were resistant to four different classes of antibiotics, while 6.9% (20/287) exhibited resistance to eight classes. The most frequent MDR pattern was PEN/CEP/TET/QNL/SUL, seen in 14.2% (38/287) of the isolates. The isolation frequency of MDR E. coli and K. pneumoniae at different sampling sites was high, being 47.6% in hospital chambers, 62.0% in hospital ponds, 58.1% in the treated hospital wastewater, and 55.6% in the community stream draining into the Indian Ocean. Extended spectrum beta-lactamase production was observed in 40% (92/230) of E. coli and 36.2% (51/141) of K. pneumoniae isolates. Resistance to quinolones among E. coli was 54.8% (126/230) and was 39.7% in K. pneumoniae (56/141). Carbapenem resistance in E. coli was 39.6% (91/230), while among K. pneumoniae isolates was 32.6% (46/141).
Conclusions
We found high proportions of multidrug-resistant E. coli and K. pneumoniae in the wastewater flowing from the hospital through the community sewage system to the point where it enters the Indian Ocean. Biological treatment did not significantly reduce the proportion of resistant bacteria, posing a very serious public health threat. The release of these highly resistant pathogens into the Indian Ocean is of international concern.
Similar content being viewed by others
Background
Antimicrobial resistance (AMR) is a global health threat and has remarkable effects on public health and the global economy (Prestinaci et al. 2015; World Health Organization 2014). Globally, it is estimated that at least 700,000 people die annually from infections that are resistant to currently available antibiotics (O’Neill 2016). Bacterial AMR is at least as large as major diseases such as HIV and malaria, with the highest burden encountered in sub-Saharan Africa (AMR Collaborators 2022). In 2019, predictive statistical models found that the highest death rates to have occurred in western sub-Saharan Africa, at 27.3 deaths per 100,000 (20.9–35.3) due to drug-resistant bacterial infections. In most African countries, AMR situation is made worse by weak regulation in antimicrobial use (AMU), tendency for animal owners to stock drugs in their houses and engaging unskilled people in treating animals (Frumence et al. 2021), limited laboratory capacity to collect and analyse data on AMR and AMU (Matee 2023), and irrational use of antibiotics in human and animal sectors, with detrimental consequences to the environmental (Fletcher 2015; Kimera et al. 2020a, b). In addition, most countries in this region do have weak health systems for AMR and AMU surveillance, crucial for production of evidence-based data needed for quantifying risks, planning, prioritization, investment of resources, inform policy development and assess the impact of intervention (Frost et al. 2021). Some sectors such as the environment are particularly underprivileged as compared to humans, animals and food of animal origin, making it difficult in implementing international guidelines at the national level (Matee et al. 2023). Indeed, a recent study suggests that sub-Saharan African countries need to fully involve clinical, veterinary and environmental departments if they are to build a robust One Health AMR preparedness response (Elton et al. 2020).
One area that deserves more attention is the role of sewage as a driver of AMR and antimicrobial resistance genes (ARGs) (Hendriksen et al. 2019). Sewage from the hospital settings serves as hotspots for AMR, where antimicrobial agent metabolites from consumed antibiotics as well as the drug-resistant bacterial pathogens in patients’ faeces and urine may be passed into the hospital sewage system (Verburg et al. 2019). As a consequence, hospital sewage often contains MDR bacteria (Zagui et al. 2020; Auguet et al. 2017) that spread rapidly into the environment by horizontal gene transfer through plasmids and transposons (Korzeniewska and Harnisz 2013). Moreover, the problem worsens as untreated or inadequately treated hospital wastewater is dumped into local sewage systems (Hocquet et al. 2016; Pärnänen et al. 2019). The effect of hospital wastewater treatment on AMR varies between studies, probably reflecting the heterogeneity of approaches used (Buelow et al. 2018).
It has been shown that AMR rates in the bacteria isolated from wastewater correlate positively with the frequency of the antibiotic resistance in the corresponding human population (Reinthaler et al. 2013). In one study, a possible transmission route for ampicillin- and ciprofloxacin-resistant Enterococcus faecium was traced from patients in hospital to urban sewage, further through wastewater treatment plants to surface water and back to humans (Iversen et al. 2004).
Gram-negative bacteria that are extended spectrum β-lactamase (ESBL) producers are of particular significance, causing infections that are particularly difficult to treat (Holmes et al. 2016). These bacteria produce a group of β-lactamases, which share the ability to hydrolyse penicillins, first-, second-, and third-generation cephalosporins, aztreonam and carry genes encoding resistance to other drug classes such as aminoglycosides (Holmes et al. 2016; Castanheira et al. 2021). E. coli and K. pneumoniae, which are also indicator bacteria in AMR surveillance in the environment (Anjum et al. 2021), represent the commonest multidrug-resistant (MDR) pathogens that exhibit ESBL, carbapenems and quinolone resistance (Castanheira et al. 2021; Cheng et al. 2018; Harmon et al. 2019; Klein et al. 2018).
In the present study, we analysed AMR patterns of E. coli and K. pneumoniae in wastewater samples collected from a regional referral hospital and along the community sewage system of Temeke District, Tanzania, up to the point of discharge in the Indian Ocean. The aim of this study was to compare the antibiotic resistance levels at the different sites of the sewage system, as well as before and after the treatment plant. This was done in order to improve our knowledge on the potential role of hospital sewage as a driver of AMR spread in the community and to determine the extent to which the treatment plan reduces antibiotic resistant bacteria. We recognize that wastewater-based epidemiology (WBE) of AMR is an important epidemiological approach to generate information on potential risk to human populations on a community scale (Choi et al. 2018). Conducting WBE on AMR by simultaneously sampling both healthcare- and community-associated sewage is gaining traction and is important in addressing the burden of AMR (Fahrenfeld and Bisceglia 2016).
Methods
Study setting
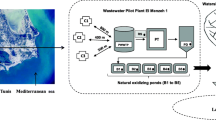
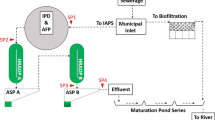
This cross-sectional study was conducted between October 2021 and January 2022 in Temeke municipality, one of the five districts in Dar es Salaam, Tanzania. The study involved microbiological examination of sewage starting from the Temeke Regional Referral Hospital (which has a bed capacity of 304, and serves more than 1,368,881 people), through various points along the community sewage system in Temeke municipality till the discharging point into the Indian Ocean (see Fig. 1).
Sampling frame and strategy
The sampling frame consisted of wastewater samples collected from wards, clinics and other administrative blocks in the hospital, as well as households, industries, markets and government institutions in the community. The samples were purposively collected from 33 sampling sites (13 from the hospital settings, 7 from the community stream and 13 from the four community ponds at Kurasini). A proportional probability-to-size sampling technique was applied to determine the number of samples to be collected from each site. A total of 400 samples were collected as follows: 109 from the hospital chambers, 48 from the hospital ponds, 85 from the community stream and 158 from the community ponds.
Sample collection and processing
At each sampling site, 3 mL of wastewater was collected using a sterile 50-mL falcon tube (BD, Nairobi, Kenya), twice a week (Monday and Thursday) for a period of 6 weeks. At the hospital, the samples were directly drawn from each inspection chamber and the general collecting chamber, whereas pond samples were taken from inlets and outlets, and samples from open streams were collected at the beginning, midpoint and end of the stream. All the samples were collected in the morning between 8:00 am and 10:00 a.m., placed in a zip-top bag, transported in a cool box at 2–8 °C, and processed in the Microbiology teaching laboratory of the Muhimbili University of Health and Allied Sciences (MUHAS) within 6 h of collection.
Isolation and identification of enteric bacteria
In the laboratory, the wastewater samples were mixed with sterile 0.9% normal saline at a ratio of 1:1 to reduce bacterial density as described by Moremi et al (2016). Then, a loopful of the diluted sample was inoculated onto MacConkey agar (Oxoid, Basingtoke, UK) and incubated aerobically at 37 °C for 18–24 h. Lactose fermenters were further examined using Gram stain and biochemical tests (Oxidase, Indole, Methyl red, Voges–Proskauer, Citrate utilization tests, and Kligler’s iron agar) for the identification of E. coli and K. pneumoniae isolates.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was performed using the Kirby Bauer disc diffusion method as per Clinical Laboratory Standard Institute (CLSI) guideline of 2019 (Clinical Laboratory Standards Institute 2019). In brief, a bacterial suspension from pure culture was adjusted to 0.5 McFarland turbidity standard, and by using a sterile cotton swab the inoculum was evenly distributed into the Mueller–Hinton agar (MHA; HiMedia Mumbai, India) and then incubated aerobically at 37 °C for 16 to 18 h. The tested antibiotic discs were ampicillin (AMP 10 µg; Oxoid, UK), tetracycline (TET 30 µg; Oxoid, UK), nalidixic acid (NAL 30 µg; Oxoid, UK), ciprofloxacin (CIP 5 µg; Oxoid, UK), imipenem (IMI 10 µg; Oxoid, UK), trimethoprim-sulfamethoxazole (SXT 1.25/23.5 µg; Oxoid, UK), gentamicin (GEN 10 µg; Oxoid, UK), cefotaxime (CTX 30 µg; Oxoid, UK) and chloramphenicol (CHL 30 µg; Oxoid, UK). An isolate was considered to be MDR if it showed resistance to at least three or more different classes of antibiotics (Magiorakos et al. 2012).
Screening and confirmation of ESBL production
All identified E. coli and K. pneumoniae isolates were inoculated onto MacConkey agar containing 2 µg /mL cefotaxime for screening and confirmed by a combination disc diffusion method, using cefotaxime (30 µg) alone and in combination with clavulanic acid (30 µg/10 µg) and ceftazidime (30 µg) alone and in combination with clavulanic acid (30 µg/10 µg). The zones of inhibition around the disc of cefotaxime alone and the disc of cefotaxime with clavulanic acid were measured. A difference of ≥ 5 mm between the two diameters indicated ESBL production as per 2019 CLSI guideline.
Screening for resistance to quinolones and carbapenems
The zones of inhibition for quinolones (nalidixic acid and ciprofloxacin) and carbapenems (imipenem) measured during susceptibility testing were used to determine the resistance to the two classes of antibiotics. For imipenem, a zone of < 19 mm (resistance) and 20–22 mm (intermediate). For nalidixic acid, a zone of < 13 mm (resistance), 14–18 mm (intermediate); and for ciprofloxacin, a zone of < 21 mm (resistant), 22–25 mm (intermediate). All intermediate zones were considered resistant (Clinical Laboratory Standards Institute 2019).
Quality control procedures
Media were prepared according to the manufacturer’s instructions, and sterility checks were performed. Standard K. pneumoniae (ATCC 700603) and E. coli (ATCC 25922) were used as positive and negative controls, respectively, during AST and when confirming ESBL-producing organisms.
Statistical analysis
Data were analysed using GraphPad Prism version 9. The proportion of MDR E. coli and K. pneumoniae was calculated by dividing the number of isolates that showed resistance to at least three different classes of antibiotics over the total number of tested isolates in a specific sewage source. Fisher’s exact test was used to compare the isolation frequency of antibiotic-resistant E. coli and K. pneumoniae between the hospital and community sewage isolates. A p value of < 0.05 was considered statistically significant.
Results
Distribution of Enterobacteriaceae in the hospital and community sewage
A total of 400 sewage samples were collected, yielding 517 isolates, of which 32.3% (167/517) were from the hospital sewage system and 67.7% (350/517) were from the community (Fig. 2A). Regarding bacterial species, E. coli was the most common 44.5%, (230/517), followed by K. pneumoniae at 27.3% (141/517) and 28.2% (146/517) of the isolates were other gram-negative bacteria (Fig. 2B).
Frequency of E. coli and K. pneumoniae isolates in the hospital and community sewage
The isolation frequency of E. coli was significantly higher in the hospital than in the community sewage, being 55.1% (92/167) against 39.4% (138/350), respectively p = 0.0009 (Fig. 3A). However, the isolation frequency of K. pneumoniae isolates from the hospital sewage (28.7% (48/167)) did not differ significantly from that of the community sewage 26.6% (93/350), p = 0.5994 (Fig. 3B).
Resistance patterns of E. coli and K. pneumoniae isolates from the hospital and community sewage
E. coli isolated from hospital and community samples were similarly highly resistant to most antibiotics except chloramphenicol, with hospital E. coli 6.5% (6/92) isolates being significantly more susceptible to chloramphenicol compared to community isolates, 26.1% (36/138), p = 0.0001. However, K. pneumoniae isolates from the hospital sewage were significantly more resistant to cefotaxime 31/48 (64.6%), against 45/93 (48.4%), gentamicin 39.6% (19/48) against 12.9% (12/93), imipenem 43.8% (21/48) against 26.9% (25/93) and sulfamethoxazole/trimethoprim 62.5% (30/48), against 36.6% (34/93) than those from the community, p < 0.05 (Table 1).
The percentage of multidrug resistance of E. coli and K. pneumoniae to tested antibiotics at different sampling points
The isolation frequency of MDR enteric bacteria at different sampling sites is shown in Fig. 4. The percentages were 47.6% in hospital chambers, 62.0% in hospital ponds, 58.1% in the treated hospital wastewater and 55.6% in the community stream draining into the Indian Ocean.
Proportion of MDR E. coli and K. pneumoniae in the hospital and community sewage
Overall, the proportion of MDR isolates was high for both E. coli 80.9% (186/230) and K. pneumoniae 71.6% (101/141) (Fig. 5A). There was no significant difference between the two sewage sources, with the frequency of hospital and community MDR E. coli being 83.7% (77/92) and 79.0% (109/138), respectively (p = 0.3979) (Fig. 5B). A similar pattern was observed among K. pneumoniae isolates, with the proportion of MDR in the hospital and community sewage isolates being 79.2% (38/48) and 67.7% (63/93), respectively, p = 0.1725 (Fig. 5C).
The patterns of MDR isolates
Overall, most of the MDR isolates were resistant to four classes of antibiotics 27.2% (78/287), and 6.9% (20/287) of the isolates were resistant to eight classes of antibiotics. The most frequent MDR pattern was PEN/CEP/TET/QNL/SUL, exhibited by 14.2% (38/287) of the isolates (Table 2).
The proportion of ESBL-producing E. coli and K. pneumoniae in the sewage
ESBL production was observed in 40% (92/230) of the E. coli isolates and 36.2% (51/141) of the isolated K. pneumoniae (Fig. 6A). The proportion of ESBL-producing E. coli was significantly higher in the community 46.4% (64/138) than in the hospital sewage 30.4% (28/92), p = 0.0194 (Fig. 6B). However, for K. pneumoniae, isolates from the hospital sewage had a significantly higher proportion of ESBL producers than those from the community, 50% (24/48) against 29% (27/93), respectively, p = 0.0168 (Fig. 6C).
Resistance patterns of ESBL-producing E. coli and K. pneumoniae from the sewage
ESBL-producing E. coli from both the hospital and community wastewater were highly resistant to most antibiotics, with hospital isolates being significantly more resistant to tetracycline than community isolates, p = 0.022. All hospital isolates of ESBL-producing E. coli were susceptible to chloramphenicol. ESBL-producing K. pneumoniae were also highly resistant to the tested antibiotics, with no significant difference seen between isolates from the two sources of wastewater (Table 3).
The proportion of quinolone-resistant E. coli and K. pneumoniae in the hospital and community sewage
Resistance to quinolones among E. coli and K. pneumoniae isolates was 54.8% (126/230) and 39.7% (56/141), respectively (Fig. 7A). There was no significant difference between community and hospital E. coli isolates, 57.2% (79/138) against 51.1% (47/92), respectively p = 0.4174 (Fig. 7B). Similarly, for K. pneumoniae, no significant differences in isolation frequency were observed between hospital sewage isolates 47.9% (23/48) and those from the community, 35.5% (33/93), p = 0.2034 (Fig. 7C).
Resistance patterns of quinolone-resistant E. coli and K. pneumoniae in hospital and community sewage
Quinolone-resistant E. coli from the hospital wastewater were more susceptible to chloramphenicol than those from the community, p = 0.0001. All quinolone-resistant K. pneumoniae from the community wastewater were susceptible to gentamycin (Table 4).
The proportion of carbapenemase-producing E. coli and K. pneumoniae in the hospital and community sewage
Carbapenem resistance among E. coli was 39.6% (91/230), while in K. pneumoniae was 32.6% (46/141) (Fig. 8A). There was no significant difference in carbapenem resistance between E. coli isolates from the hospital 41.3% (38/92) and those from the community 38.4% (53/138), p = 0.6815 (Fig. 8B). Similarly, the proportion of hospital K. pneumoniae isolates with carbapenem resistance 43.8% (21/48) did not significantly differ from that of the community sewage 26.9% (25/93), p = 0.0577 (Fig. 8C).
Resistance patterns of carbapenemase-producing E. coli and K. pneumoniae in the hospital and community sewage
Carbapenem-resistant E. coli from the hospital sewage were significantly less resistant to chloramphenicol compared to community isolates. Additionally, they were highly resistant to gentamycin and sulfamethoxazole/trimethoprim like their community counterparts. Carbapenem-resistant K. pneumoniae from the hospital sewage were significantly more resistant to gentamicin and sulfamethoxazole/ trimethoprim than the community isolates (Table 5).
Discussion
In this study, we found high proportions of drug-resistant, including MDR E. coli and K. pneumoniae, in the wastewater samples from all sampled sites from the hospital through the community sewage system to the point where it enters the Indian Ocean. A quarter of the isolates showed resistance to four antimicrobial classes, while some of the isolates resisted all eight classes of antimicrobial drugs. The most frequent MDR pattern was PEN/CEP/TET/QNL/SUL, exhibited by most of the isolates. In addition, biological treatment did not significantly reduce the proportion of resistant bacteria isolated from the hospital ponds compared to those isolated from hospital effluents. We found some variations in the proportion of quinolone and carbapenem-resistant E. coli and K. pneumoniae isolated from the hospital and community sewage wastewater. The isolation frequency of quinolone-resistant E. coli was higher in the community wastewater (57.3%) than among the hospital isolates (51.7%). Comparable findings were reported in Romania, whereby the proportions of MDR E. coli from the hospital and community wastewater were 85.11% and 73.53%, respectively (Gaşpar et al. 2021). Another study conducted in Nigeria found that hospital sewage harboured a high proportion (86.9%) of MDR E. coli and K. pneumoniae, with the majority showing resistance to more than two classes of tested antibiotics (Osadebe and Okounim 2020). We found co-occurrence of ESBL and carbapenemase resistance in many MDR E. coli and K. pneumoniae isolates, due to shared transfer mechanisms, implying that infections with such bacteria often result in high morbidity and mortality rates (Mazzariol et al. 2017).
Our results have several implications: (i) the persistence of a high proportion of MDR bacteria certainly indicates that the sewage system in the studied area is a major driver of AMR in the community, (ii) the lack of significant difference in reduction in the trend of isolation frequencies of MDR bacteria from the hospital pond (62.0%) to 58.1% from the treated effluent released to the community streams and 58.6% in the community sewage wastewater before being released to the Indian ocean signifies ineffective biological treatment of sewage wastewater, at the treatment plants, and (iii) the loads of highly resistant bacteria being discharged into Indian Ocean poses public health issues of international concern. This is highly significant given the fact the World Health Organization has classified Enterobacteriaceae, carbapenem-resistant, and ESBL-producing bacteria as critical pathogens that can pass along genetic material that allows other bacteria to become resistant to the best available antibiotics for treating MDR bacteria (WHO 2017). The classification was based on how deadly the infections they cause are, specifically (i) duration of hospital stays, (ii) frequency of potential occurrence of resistance to existing antibiotics, (iii) the extent of spread between animals, from animals to humans, and (iv) whether new antibiotics to treat them are already in the research and development (R&D) pipeline.
Our findings conform to other studies that have shown hospital wastewater to be the hotspot for ARB and ARG, especially high proportion of MDR-E. coli with the potential of being transmitted to the community (Gumede et al. 2021). The lack of appreciable reduction in ARB and ARG in the wastewater treatment plant effluents has also been shown by others (Leclercq et al. 2013; Okoh and Igbinosa 2010) leading to large amounts of resistant bacteria, of hospital origin being released into the recipient waters (Rizzo et al. 2013a, b). In Temeke, waster is treated biologically by aerobic digestion technique, which does not seem to be effective. During the study, we observed the following constraints in sewage management practices; low priority accorded to sanitation and hygiene improvement, inadequate investment financial resources, fragmented planning, limited participation of beneficiaries and other stakeholders, inadequate availability of effective sewerage and sanitation systems, lack of attention on selecting the most appropriate technology and general low public awareness. This calls for significant allocation of resources and modification in wastewater treatment protocols and continuous monitoring for AMR and antimicrobial groups (AMG). The study of resistant microbes in sewage should cover a range of factors, including the evolution of resistance at the molecular level within a given organism, transmission mechanisms and pathways between organisms, and dissemination to humans and animal hosts and across the wider environment including soil and water. Indeed, hospital and community wastewater are now known to be the source of AMR transmission within the environment (Daoud et al. 2017), and therefore, curbing of AMR needs to involve a One Health approach since all three compartments need to be considered (O’neill 2016). We advocate that future research on AMR and sewage should focus on identifying the influence of various interventional activities such as (i) antibiotic stewardship in hospital settings, environmental sanitation (effective disposal of waste), transmission pathways-resistant bacteria and cost-effective sustainable technological, social and economic initiatives for the mitigation of environmental antibiotic resistance.
Although this study has provided valuable information to the international community, we do acknowledge some limitations. First, the study was conducted in dry season, the pattern of AMR could have seasonal variations (Ramsey et al. 2019). Secondly, due to logistical issues we could not perform whole genome sequencing (WGS) of the MDR E. coli and K. pneumoniae, which could have provided an insight into the AMR transmission dynamics of the wastewater from the hospital sewage down the stream to the Indian Ocean. Future studies should use advanced technologies such as WGS and metagenomics and involve several compartments (sewage and the surrounding community) to decipher dynamics and transmission patterns and pathways of resistomes among the various compartments. This approach is important given the immense diversity of antibiotic resistance genes (ARGs), the complexity of ARG transfer, and the broad range of omnipresent factors contributing to AMR.
Conclusions
The high proportions of drug-resistant, including multidrug-resistant, E. coli and K. pneumoniae in the wastewater samples from all sampled sites from the hospital through the community sewage system to the point where it enters the Indian Ocean are a major threat to public and the environment. The lack of significant difference in reduction in the trend of isolation frequencies of MDR bacteria from the hospital sewage system to the Indian Ocean signifies ineffective biological treatment of sewage wastewater, at the treatment plants. The loads of highly resistant bacteria being discharged into Indian Ocean possess public health issues of international concern. Strict implementation of appropriate disinfection technologies for hospital sewage would reduce the bacterial load in the sewage that will reach urban wastewater treatment plants, minimizing the spread of the resistance pathogens in the environment.
Availability of data and materials
All data generated and/ or analysed during this study are included in this published article.
Abbreviations
- AMP:
-
Ampicillin
- AMR:
-
Antimicrobial resistance
- AMU:
-
Antimicrobial use
- ARB:
-
Antibiotic-resistant bacteria
- ARG:
-
Antibiotic resistance genes
- ATCC:
-
American Type Culture Collection
- CHL:
-
Chloramphenicol
- CIP:
-
Ciprofloxacin
- CLSI:
-
Clinical Laboratory Standards Institute
- CTX:
-
Cefotaxime
- DMDP:
-
Dar es Salaam Metropolitan Development Project
- E. coli :
-
Escherichia coli
- ESBL:
-
Extended spectrum beta lactamase
- GEN:
-
Gentamicin
- IMI:
-
Imipenem
- IPC:
-
Infection prevention and control
- K. pneumoniae :
-
Klebsiella pneumoniae
- KCMC:
-
Kilimanjaro Christian Medical College
- KIA:
-
Kligler Iron Agar
- MDR:
-
Multidrug-resistant
- MHA:
-
Mueller Hinton Agar
- MUHAS:
-
Muhimbili University of Health and Allied Sciences
- NAL:
-
Nalidixic acid
- SPSS:
-
Statistical Package for Social Sciences
- SXT:
-
Sulphamethoxazole/trimethoprim
- TET:
-
Tetracycline
- TRRH:
-
Temeke Regional Referral Hospital
- UK:
-
United Kingdom
References
AMR Collaborators (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399(10325):629–655. https://doi:https://doi.org/10.1016/S0140-6736(21)02724-0
Anjum MF, Schmitt H, Börjesson S, Berendonk TU (2021) The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. https://doi.org/10.1016/j.mib.2021.09.011
Auguet O, Pijuan M, Borrego CM, Rodriguez-Mozaz S et al (2017) Sewers as potential reservoirs of antibiotic resistance. Sci Total Environ 605–606:1047–1054. https://doi.org/10.1016/j.scitotenv.2017.06.153
Buelow E, Bayjanov JR, Majoor E, Willems RJ, Bonten MJ, Schmitt H, van Schaik W (2018) Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol Ecol 94(7)
Castanheira M, Simner PJ, Bradford PA (2021) Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrob Resist 3(3). https://doi.org/10.1093/jacamr/dlab092
Cheng VCC, Wong SC, Wong SCY, Ho PL, Yuen KY (2018) Control of carbapenemase-producing enterobacteriaceae: beyond the hospital. E Clin Med 6:3–4. https://doi.org/10.1016/j.eclinm.2018.12.008
Choi P, Tscharke B, Donner E, O’Brien J, Grant S, Kaserzon S, Mackie R, O’Malley E, Crosbie N, Thomas K, Mueller J (2018) Wastewater-based epidemiology biomarkers: past, present and future. Trends Anal Chem 105:453–469. https://doi.org/10.1016/j.trac.2018.06.004
Clinical Laboratory Standards Institute (2019) M100-S11, Performance standards for antimicrobial susceptibility testing. Clin Microbiol Newsletter 23(6):49. https://doi.org/10.1016/s0196-4399(01)88009-0
Daoud Z, Farah J, Salem ES, Kfoury KEl, Dahdouh E (2017) Multidrug-resistant enterobacteriaceae in lebanese hospital wastewater: implication in the one health concept. Microb Drug Resist. https://doi.org/10.1089/mdr.2017.0090
Elton L, Thomason MJ, Tembo J, Velavan TP, Pallrla SG, Arunda LB et al (2020) Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob Resist Infect Control 9:145. https://doi.org/10.1186/s13756-020-00800-y
Fahrenfeld N, Bisceglia KJ (2016) Emerging investigators series: Sewer surveillance for monitoring antibiotic use and prevalence of antibiotic resistance: urban sewer epidemiology. Environ Sci Water Res Technol 2:788–799. https://doi.org/10.1039/c6ew00158k
Fletcher S (2015) Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ Health Prev Med 20(4):243–252. https://doi.org/10.1007/s12199-015-0468-0
Frost I, Kapoor G, Craig J, Liu D, Ramanan Laxminarayan R (2021) Status, challenges and gaps in antimicrobial resistance surveillance around the world. J Glob Antimicrob Resist. https://doi.org/10.1016/j.jgar.2021.03.016
Frumence G, Mboera LEG, Sindato C, Durrance-Bagale A, Jung A, Mshana SE, Clark TG, Legido-Quigley H, Matee MI (2021) Practices and Challenges of Veterinary Paraprofessionals in Regards to Antimicrobial Use and Resistance in Animals in Dar Es Salaam, Tanzania. Antibiotics 10:733
Gaşpar CM, Cziszter LT, Lăzărescu CF, Ţibru I, Pentea M, Butnariu M (2021) Antibiotic resistance among Escherichia coli isolates from hospital wastewater compared to community wastewater. Water 13(3449):1–11. https://doi.org/10.3390/w13233449
Gumede SN, Abia ALK, Amoako DG, Essack SY (2021) Analysis of wastewater reveals the spread of diverse extended-spectrum β-lactamase-producing E. coli strains in uMgungundlovu District , South Africa. 1–15.
Harmon DE, Miranda OA, McCarley A, Eshaghian M, Carlson N, Ruiz C (2019) Prevalence and characterization of carbapenem-resistant bacteria in water bodies in the Los Angeles-Southern California area. Microbiol Open 8(4):1–13. https://doi.org/10.1002/mbo3.692
Hendriksen RS, Munk P, Njage P et al (2019) Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun 10:1124. https://doi.org/10.1038/s41467-019-08853-3
Hocquet D, Muller A, Bertrand X (2016) What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93(4):395–402. https://doi.org/10.1016/j.jhin.2016.01.010
Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, Piddock LJ (2016) Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387(10014):176-187
Iversen A, Kühn I, Rahman M, Franklin A, Burman LG, Olsson-Liljequist B, Torell E, Mollby R (2004) Evidence for transmission between humans and the environment of a nosocomial strain of Enterococcus faecium. Environ Microbiol 6:55–59. https//doi:https://doi.org/10.1046/j.1462-2920.2003.00534.x
Kimera ZI, Frumence G, Mboera LEG, Rweyemamu M, Mshana SE, Matee MIN (2020a) Assessment of drivers of antimicrobial use and resistance in poultry and domestic pig farming in the Msimbazi river basin in Tanzania. Antibiotics 9(12):1–20. https://doi.org/10.3390/antibiotics9120838
Kimera ZI, Mshana SE, Rweyemamu MM, Mboera LEG, Matee MIN (2020b) Antimicrobial use and resistance in food-producing animals and the environment: an African perspective. Antimicrob Resist Infect Control 9(1). https://doi.org/10.1186/s13756-020-0697-x
Klein EY, van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci USA 115(15):E3463–E3470. https://doi.org/10.1073/pnas.1717295115
Korzeniewska E, Harnisz M, (2013) Beta-lactamase- producing Enterobacteriaceae in hospital effluents. J Environ Manag 123:1–7. http://dx.doi.org/https://doi.org/10.1016/j.jenvman.2013.03.024
Leclercq R, Oberle K, Galopin S, Cattoir V, Budzinski H, Petit F (2013) Changes in Enterococcal populations and related antibiotic resistance along a medical center–wastewater treatment plant–river continuum. Appl Environ Microbiol 2013 (79):2428–2434. https//doi:https://doi.org/10.1128/AEM.03586-12
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Matee M, Mshana SE, Mtebe M et al (2023) (2023) Mapping and gap analysis on antimicrobial resistance surveillance systems in Kenya, Tanzania, Uganda and Zambia. Bull Natl Res Cent 47:12. https://doi.org/10.1186/s42269-023-00986-2
Mazzariol A, Bazaj A, Cornaglia G (2017) Multi- drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemotherapy 29:2–9. https://doi.org/10.1080/1120009X.2017.1380395
Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, Chakraborty T, Mshana SE (2016) Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front Microbiol, https://doi.org/10.3389/fmicb.2016.01862
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D (2013a) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447,345–360. https// doi:https://doi.org/10.1016/j.scitotenv.2013.01.032
Okoh AI, Igbinosa EO (2010) Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol 10:143. https//doi:https://doi.org/10.1186/1471-2180-10-143
O’Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf
O’neill J (2016) Tackling drug-resistant infections globally: an overview of our work the review on antimicrobial resistance
Osadebe AU, Okounim B (2020) Multidrug-resistant bacteria in the wastewater of the hospitals in Port Harcourt metropolis: implications for environmental health. https://doi.org/10.22102/jaehr
Pärnänen KM, Narciso-da-Rocha C, Kneis D, Berendonk TU, Cacace D, Do TT, Manaia CM et al (2019) Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci Adv 5(3)
Prestinaci F, Pezzotti P, Pantosti A (2015) Antimicrobial resistance: a global multifaceted phenomenon. Pathogens Global Health 109(7):309–318. https://doi.org/10.1179/2047773215Y.0000000030
Ramsey EG, Royer J, Bookstaver PB, Justo JA, Kohn J, Albrecht H, Al-Hasan MN (2019) Seasonal variation in antimicrobial resistance rates of community-acquired Escherichia coli bloodstream isolates. Int J Antimicrob Agents 54(1):1–7. https://doi.org/10.1016/j.ijantimicag.2019.03.010. (Epub 2019 Mar 15)
Reinthaler FF, Galler H, Feierl G, Haas D, Leitner E, Mascher F, Melkes A, Posch J, Pertschy B, Winter I, et al (2013) Resistance patterns of Escherichia coli isolated from sewage sludge in comparison with those isolated from human patients in 2000 and 2009. J. Water Health 11:13–20. https//doi:https://doi.org/10.2166/wh.2012.207
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D (2013b) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. https://doi.org/10.1016/j.scitotenv.2013.01.032
Verburg I, García-Cobos S, Hernández Leal L, Waar K, Friedrich AW, Schmitt H (2019) Abundance and antimicrobial resistance of three bacterial species along a complete wastewater pathway. Microorganisms 7(9):312
World Health Organization (2014) Global report on surveillance 2014. WHO 2014 AMR Report, pp 1–8
WHO (2017) Media Centre. News Release. WHO publishes list of bacteria for which new antibiotics are urgently needed. 2017 [cited 2017 March]. Available from: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/
Zagui GS, Andrade LN, Moreira NC, Silva TV, Machado GP, Darini ALC, Segura-Muñoz SI (2020) Gram-negative bacteria carrying β-lactamase encoding genes in hospital and urban wastewater in Brazil. Environ Monitor Assessment 192(6):376. https://doi.org/10.1007/s10661-020-08319-w
Acknowledgements
We firmly acknowledge the cooperation we received from Temeke Regional Referral Hospital (TRRH) (Environment health office)), Temeke Municipal Council, and the Dar es Salaam Water Supply and Sanitation Authority (DAWASA, Kurasini ponds).
Funding
This study did not receive any external funding.
Author information
Authors and Affiliations
Contributions
NZS, MIM and FM contributed to conception and study design; NZS, ZK and FXM contributed to data collection and laboratory investigations; NZS, MIM and FM analysed the data; AM, NZS, FM and MIM drafted the initial manuscript; AM, NZS, ZK, FM, AJ and MIM reviewed the final manuscript. All the authors read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Research and Publications Committee of the Muhimbili University of Health and Allied Sciences (MUHAS) approved the study reference number DA.282/298/01.C/. Permission to conduct the study was granted by Temeke regional referral hospital and the Dar es Salaam water supply and sanitation authority (DAWASA).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Seguni, N.Z., Kimera, Z.I., Msafiri, F. et al. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from hospital sewage flowing through community sewage system and discharging into the Indian Ocean. Bull Natl Res Cent 47, 66 (2023). https://doi.org/10.1186/s42269-023-01039-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-023-01039-4