Abstract
Background
In 2003, the first case of severe acute respiratory syndrome coronavirus (SARS-CoV) was recorded. Coronaviruses (CoVs) have caused a major outbreak of human fatal pneumonia. Currently, there is no specific drug or treatment for diseases caused by SARS CoV 2. Computational approach that adopts dynamic models is widely accepted as indispensable tool in drug design but yet to be exploited in covid-19 in Zaria, Nigeria. In this study, steps were taken to advance on the successful achievements in the field of covid-19 drug, with the aid of in silico drug design technique, to create novel inhibitor drug candidates with better activity. In this study, one thousand human immunodeficiency virus (HIV1) antiviral chemical compounds from www.bindingBD.org were docked on the SARS CoV 2 main protease protein data bank identification number 6XBH (PDB ID: 6XBH) and the molecular docking score were ranked in order to identify the compounds with the highest inhibitory effects, and easy selection for future studies.
Results
The docking studies showed some interesting results. Inhibitors with Index numbers 331, 741, and 819 had the highest binding affinity. Similarly, inhibitors with Index number 441, 847, and 46 had the lowest hydrogen bond energy. Inhibitor with index number 331 was reported with the lowest value (− 48.38kCal/mol). Five new compounds were designed from the selected six (6) compounds with the best binding score giving a total of thirty (30) novel compounds. The low binding energy of inhibitor with index no. 847b is unique, as most of the interaction energies are of H-bond type with amino acids (Thr26, Gly143, Ser144, Cys145, Glu166, Gln189, Hie164, Met49, Thr26, Thr25, Thr190, Asn142, Met165) resulting in an overall negative value (−16.31 kCal/mol) making it the best of all the newly designed inhibitors.
Conclusions
The novel inhibitor is 2-(2-(5-amino-2-((((3-aminobenzyl)oxy)carbonyl)amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid. The improvement it has over the parent inhibitor is from the primary amine group attached to meta position of first benzene ring and the carboxyl group attached to the ortho position of the second benzene ring. The molecular dynamics studies also show that the novel inhibitor remains stable after the study. This result makes it a better drug candidate against SARS CoV 2 main protease when compared with the co-crystallized inhibitor or any of the 1000 docked inhibitors.
Similar content being viewed by others
Background
Coronaviruses (CoVs) have caused a major outbreak of human fatal pneumonia since the beginning of the twenty-first century. COVID‐19 is an acute respiratory disease caused by the RNA virus SARS‐CoV‐2. In severe cases, the infection can cause pneumonia, severe acute respiratory syndrome, kidney failure, and even death (Du et al. 2020). There is currently no specific medicine or treatment for diseases caused by SARS-CoV-2 (Huang et al. 2020).
SARS-CoV-2 virus targets cells through the viral structural spike (S) protein that binds to the angiotensin-converting enzyme 2 (ACE2) receptor. Once inside the cell, viral polyproteins are synthesized that encode for the replicase-transcriptase complex. Structural proteins are synthesized leading to completion of assembly and release of viral particles (De Groot et al. 2013; Lau et al. 2005; Reusken et al. 2013).
Hosseini and Amanlou (2020) conducted a virtual screening procedure employing docking of 1615 FDA approved drugs to identify new potential small molecule inhibitors for protease protein of COVID-19 and their result indicates that among all FDA-approved drugs, simeprevir which is used for Hepatitis C virus (HCV) NS3/4A protease inhibitor, revealed strong interaction with protease binding pocket and placed well into the pocket even better than the lopinavir-ritonavir (Abd El-Aal et al. 2022; Al-Hossainy et al. 2021; El Azab et al. 2021). Since this compound is FDA-approved and has successfully passed various testing steps, they suggested that this drug could be a potential drug for treating the COVID-19 (Hosseini & Amanlou 2020).
Motiwale et al. (2020) and friends applied molecular docking approach in conjugation with molecular dynamics (MD) simulations to find out potential inhibitors against Mpro of SARS-CoV-2 from previously reported SARS-3CL protease inhibitors. They used a total of 61, previously known inhibitors, where 4-(Sacco et al. 2020) benzoic acid, and 4-(4-methoxyphenyl)-6-oxo-2-[(2-phenylethyl)sulfanyl]-1,6-dihydropyrimidine-5-carbonitrile were reported to have minimum and maximum binding energy, respectively (Motiwale et al. 2020).
To achieve a fast and reliable drug in this current crisis, we initiated a virtual screening procedure(Gagic et al. 2020), employing docking of 1000 HIV1 protease inhibitors compounds from www.bindingBD.org over binding pocket of SARS CoV 2 main protease using 1 pdb file PDB: (6XBH) downloaded from Research Collaboratory for Structural Bioinformatics (RCSB) which represent main protease of SARS CoV 2 to identify potent inhibitors against the virus and to design a novel drug whose molecular dynamics studies will be done to ascertain the effectiveness in inhibiting the SARS-CoV-2 Mpro (Sacco et al. 2020).
The molecular docking result would provide first-hand knowledge about the interactions between the ligands and the target receptors since most of these ligands work by profoundly inhibiting the specificity and efficiency of protein (target) action (Adeniji et al. 2020; Stockwell 2000). All these will undoubtedly offer important structural insight into the design of novel COVID-19 drugs (Abd El-Aal et al. 2022; Al-Hossainy et al. 2021; El Azab et al. 2021). Besides, these methods will help to: promote savings in the cost of drug design and development, reduce the requirement for lengthy and expensive animal tests and, promote green chemistry to increase efficiency and eliminate chemical waste (DiMasi et al. 2003). Overall, the purpose of the work is to carry out a computer-aided drug design (CADD) of SARS CoV 2 main protease inhibitor.
Methods
Software
The following is a list of software used in this research work: Molsoft.icm-pro.v3.8.3 software built Nov 30 2014 20:23. 4.7.5. © Copyright 1989–2022, MolSoft L.L.C. 11199 Sorrento Valley Road, S209 San Diego CA 92121 (Abagyan et al. 1994), Visual Molecular Dynamics (VMD 1.9, 2011-03-14 Platforms) Windows OpenGL, CUDA (Windows XP/Vista/7/8/10 (32-bit) with OpenGL and CUDA) (Humphrey et al. 1996) and Nanoscale Molecular Dynamics (NAMD) version 2.14 (2020-08-05) Platforms Win64 software (freeware license) which were developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign." (Phillips et al. 2020), Discovery studio Client v21.1.0.2098, Copyright © 2020, Dessault Systemes Biovia Corp (Biovia 2017), Molecular Operating Environment (MOE), 2020.09 Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2022 (Environment 2014) and Spartan'14, version 1.1. 2 Wavefunction, Inc 18401 Von Karman Ave., Suite 435 Irvine, CA 92612 (Wavefunction 2013; Baig et al. 2020).
Experimental dataset
In this study, a dataset of 1000 HIV 1 antiretroviral compounds presented in Additional file 1: Table S1 was used for molecular docking and molecular dynamics studies to generate a novel inhibitor for the SARS CoV 2 main protease. These compounds are derivatives of: diazepam-2-one, benzamide, butanamide, carbamate, thiadiazepane 1,1-dioxide, thiadiazepane 1,1-dione, hexanediamide, dihydro-2H-pyran-2-one, benzenesulfonate imidazole-2-sulfonate, sulfamate,
butanediamide, chromen-2-one, sulfonamide, thiazolidine-4-carboxamide, phenylpentanamide, piperidine-2-carboxamide, benzenesulfonamide, pyridine-2-sulfonamide, pyrimidinone, coumarin, pyran-2-on.
Molecular docking study
Ligand preparation
The 2D structure of each inhibitor was drawn using the ChemDraw v16.0 Windows 10 (32 bit and 64 bit), Copyright 1998–2016 PerkinElmer Informatics Inc and presented in table (Arthur et al. 2020). The structures were introduced into wavefunction 14 graphic user interphase (GUI) after which the 2D structures were converted into 3D structures by selecting the view dialog box present on Spartan 14 GUI. From the build option on Spartan 14, the structures were clean by checking to minimize using molecular mechanic force field (MM+) option to remove all strain from the molecular structure. In addition, this will ensure a well-defined conformer relationship among compounds of the study (Viswanadhan, Ghose, Revankar, & Robins, 1989). From the setup calculation option on Spartan 14, the calculation was set to equilibrium geometry at the ground state using a semi-empirical PM6 (Parameterization Method 6) (Bikadi and Hazai 2009).
Preparation of receptor
The x-ray diffraction crystal structure SARS-CoV-2 (COVID-19) main protease with PDB ID: 6XBH (Sacco et al. 2020) with a resolution of 1.60 Å was used for the study. The complexed inhibitor, (R)-3-(((2R,5S)-5-(((S)-(benzyloxy)(hydroxy)methyl) amino)-1-hydroxy-4-oxo-6-phenyl hexan-2-yl) amino) -1,3-dihydro-2H-pyrrol-2-one was removed from the chain of 6XBH where it was covalently bonded with the DNA in the receptor.
The receptor structure was imported into the Molsoft.icm-pro.v3.8.3 GUI (Arthur and Uzairu 2019), and the PDB files were converted into an internal coordinate mechanics (ICM-object) (MolSoft, 2000) by deleting the additional water molecules confined in the X-ray structure collected from the PDB data bank. All the hydrogen atoms were optimized before the receptor was then subjected to the process of molecular docking treatment (Sastry et al. 2013).
There are five different interaction potentials that contribute to the overall free binding energy established between the receptor pocket and the docked ligand (Gallicchio et al. 2010). These potentials include van der Waals potential for a hydrogen atom probe, van der Waals potential for a heavy-atom probe (generic carbon of 1.7A radius), hydrophobic energy terms, optimized electrostatic energy term, and lone-pair-based potential, which reflects directional preferences in hydrogen bonding. These energy terms are based on the all-atom vacuum force field with added functions to account for solvation free energy, desolvation energy and entropic contribution. It was shown that after each random step, full local minimization greatly improves the efficiency of the procedure (Abd El-Aal et al. 2022; Al-Hossainy et al. 2021; Ibrahim et al. 2020; Mohamed et al. 2022; Zwawi et al. 2021). The ICM program relies on global optimization of the entire flexible ligand in the receptor field and combines large-scale random moves of several types with gradient local minimization and a search history mechanism (Arthur et al. 2018).
Molecular dynamics simulation
Molecular dynamics (MD) simulations were carried out on the 3D crystal structure of SARS-CoV-2 (COVID-19) main protease with PDB ID: 6XBH (Sacco et al. 2020) in complex with the reference inhibitor (R)-3-(((2R,5S)-5-(((S)-(benzyloxy) (hydroxy) methyl)amino)-1-hydroxy-4-oxo-6-phenylhexan-2-yl)amino)-1,3-dihydro-2H-pyrrol-2-one. The 3D crystal structure of SARS-CoV-2 (COVID-19) main protease with PDB ID: 6XBH (Sacco et al. 2020) was extracted from the crystal structure complex with reference inhibitor. This was complexed with the best inhibitor for MD simulations studies as well. MD simulations were carried out using the AMBER version 11 package with the ff99SB force field (Hornak et al. 2006).
The protein structure was surrounded with a 15 Å layer of TIP4P BOX water molecules. The electrostatic charge was neutralized by adding counter ions using the LeaP program of AMBER ver.11. After minimization, heating and equilibration, the production MD phase was carried out at 300 K for 1 ns with a time step of 1 ps (picoseconds) using the constant volume and temperature (NVT) ensemble and the Particle Mesh Ewald algorithm for the calculation of electrostatic interactions (Haspel, Zheng, Aleman, Zanuy, & Nussinov, 2017). The initial velocity of atoms was generated at 100 K in heating phase with a Maxwellian distribution and maintained. The pressure was kept at 1 bar by Berendsen weak coupling approach during equilibration (Berendsen et al. 1984).
Results
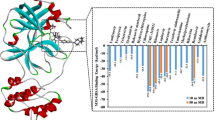
The numerical results of this study are presented in the tables presented below. This has become necessary because of the need to correlate some of the data. Other results, such as plots and pictorial representation of the interactions between the ligands and their receptor binding sites are presented as figures.
Table S2 shows the molecular docking result of the reference inhibitor and 1000 HIV 1 antiviral compounds on SARS-CoV-2 main protease receptor (PDB ID: 6XBH). The following parameters are shown: number of rotatable torsions, hydrogen bond energy, hydrophobic energy in exposing a surface to water, the van der Waals interaction energy (sum of gc and gh van der Waals), internal conformational energy of the ligand, the desolvation of exposed H-bond donors and acceptors, the solvation electrostatics energy change upon binding and mean force score. According to the molecular docking results, it was found that the binding energy of co-crystallized ligand, (R)-3-(((2R,5S)-5-(((S)-(benzyloxy)(hydroxy)methyl)amino)-1-hydroxy-4-oxo-6-phenylhexan-2-yl) amino)-1,3-dihydro-2H-pyrrol-2-one was − 23.56kCal/mol while the binding energy of all the 1000 HIV 1 antiviral inhibitors lies between − 4.73 and − 48.38 kCal/mol. Figure 1 is the docked poses of SARS CoV-2 main protease (PDB ID: 6XBH) with REF-IN (stick figure) while Table 3 shows the interaction types with surrounding amino acids of SARS CoV 2 Main Protease (PDB ID: 6XBH) with REF-IN.
REF-IN is the reference inhibitor with IUPAC name (R)-3-(((2R,5S)-5-(((S)-(benzyloxy) (hydroxy) methyl) amino)-1-hydroxy-4-oxo-6-phenylhexan-2-yl) amino)-1,3-dihydro-2H-pyrrol-2-one.
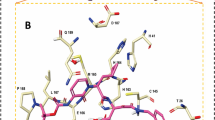
REF-IN binds at the ligand binding site and forms seven (7) H-bonds with critical amino acids residue in the ligand binding domain site of 6XBH, four (4) of which are conventional H-bonds involving Asn142, Ser144, Gln189, His163 and three carbon-hydrogen bonds between the ligand and Leu141, Glu166 and His163 which are clearly shown in Fig. 1. Other interactions are Π-Pi stacked with Hie41, Π-sulfur interaction with Met165 and alkyl-type interaction with Met49 and Met165 shown in Table 1. The ICM score for the best possible interaction pose was given as − 23.56 kCal/mol.
The molecular docking score was ranked in order to identify the compounds with the highest inhibitory effects, and easy selection for future studies. The docking studies showed that inhibitors with Index numbers 331, 741, and 819 had the highest binding energies of all the compounds that were docked on SARS CoV 2 main protease (Additional file 1: Table S2). Similarly, inhibitors with Index number 441, 847, and 46 had the highest hydrogen bond energy. Inhibitor with index number 331 was reported with the highest value (− 48.38 kCal/mol) and inhibitor with index number 46 having the least value (− 15.69 kCal/mol) for all the best six (6) selected. The high correlation of H-bonds with the number of flexible bonds (nflex) reflects on the high binding energies of the compounds, with the exception of inhibitor with index number 331 which has six (6) flexible bonds. The low binding energy of inhibitor with index number 331 is unique, its amplified hydrogen bond energy was as a result of an inductive effect created by the presence of three Π-sulfur interactions observed with Cys145, Met165 and Cys145, an amide pi stacked interactions with Thr24 and Thr25, π-lone pair interaction with Thr24, π-pi stacked interaction with Hie41 and π-alkyl interaction involving Met49 and Cys145. Another noticeable point is the bond length of two of the conventional H-bonds involving Thr26 and Hie164 with bond length 1.68 and 1.94A, respectively, thus impacting positively on its activity.
Based on binding energy ranking, inhibitors with Index numbers 331, 741, and 819 were selected for design. Similarly, inhibitors with Index numbers 441, 847, and 46 were selected based on hydrogen bond energy for design. Five new inhibitors labeled a-e were designed for each of the selected inhibitors above. All the compounds in the dataset docked were found to inhibit the receptor by completely occupying the active sites in the target receptor. Most of the inhibitors were tangled in both hydrophobic and hydrogen bonding interactions with the receptor. Here, it was found that strong inhibitor binding is reflected by the frequency of hydrogen bonds. In addition, the molecular docking studies carried out show that all the compounds were found to inhibit the receptors by completely occupying the active sites in the target receptor. The mechanism for this reaction is the same in all cases, which includes the intercalation of the inhibitors between the covalently bonded SARS CoV 2 main protease complex. Additional file 1: Table S3 shows the structures and IUPAC name of designed novel inhibitors while Table 8 shows the molecular docking results. From the table of docking studies, inhibitors with Index numbers 741a, 847b and 741d had the highest binding energies of all the compounds that were docked on SARS CoV 2 main protease. Similarly, inhibitors with Index numbers 847b, and 46d had hydrogen bond energy of − 16.31 kCal/mol and 15.69 kCal/mol, respectively. Inhibitor with index number 741a was reported with the lowest binding energy value of − 45.33 kCal/mol and inhibitor with index number 46d have the least binding energy value of − 34.35 kCal/mol for all the best four (4) selected designed novel inhibitors. The high binding energy of inhibitor with index number 847b is unique, as most of the interaction energies are of H-bond type with amino acids (Thr26, Gly143, Ser144, Cys145, Glu166, Gln189, Hie164, Met49, Thr26, Thr25, Thr190, Asn142, Met165) resulting in an overall negative value. The result could be partly explained by the fact that the inhibitor has nineteen (19) hydrogen bond interaction with the amino acids of the binding pocket of the SARS CoV 2 main protease which is evidenced by the high hydrogen bond energy value of − 16.31 kCal/mol making it the highest of all the newly designed inhibitors. Other noticeable interactions with the receptor include π-alkyl interaction mediated through Cys145. The inhibitor benzyl (5-amino-1-((4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxy-1-phenylbutan-2-yl)amino) -1,5-dioxopentan-2-yl) carbamate (Index number 847) from which it was designed has binding score energy of − 39.89 kCal/mol and H-bond energy of − 10.27 kCal/mol as against binding score energy and hydrogen bond energy of − 41.32 and − 16.31 kCal/mol, respectively, for the novel inhibitor. The molecular dynamics studies also show that all the hydrogen bonds formed by the compound with index number 847b remain stable after the study.
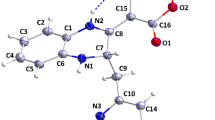
2-(2-(5-amino-2-((((3-aminobenzyl)oxy)carbonyl) amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid differ significantly in activity from its parent chain because of the introduction of primary amine group attached to meta position of first benzene ring and the carboxyl group attached to the ortho position of the second benzene ring. These groups have the ability to increase the overall binding energy by increasing the number of hydrogen bonds interaction present in their complex. This effects makes 2-(2-(5-amino-2-((((3-aminobenzyl)oxy)carbonyl) amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino)butanamido)-3-hydroxybutyl) benzoic acid a better drug candidate against SARS CoV 2 main protease with the binding energy of -41.32 kCal/mol.
The docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) and inhibitors with Index numbers 331, 441, 46, 741, 819, 847 are presented in Figs. 2, 3,4, 5, 6 and 7, while Tables 2, 3, 4, 5, 6 and 7 show the interaction types with surrounding amino acids of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor Index numbers 331, 441, 46, 741, 819, 847.
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 331 (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 331; b 3D view of inhibitor with Index number 331 with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 331 with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 441 (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 441; b 3D view of inhibitor with Index number 441 with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 441 with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 46 (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index no. 46; b 3D view of inhibitor with Index number 46 with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 46 with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 741 (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 741; b 3D view of inhibitor with Index number 741 with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 741 with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 819 (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 819; b 3D view of inhibitor with Index number 819 with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 819 with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 847 (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 847; b 3D view of inhibitor with Index number 847 with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 847 with surrounding amino acids of 6XBH
Table S3 presents the structure and IUPAC name of designed novel inhibitors, while Table 8 presents the molecular docking result of designed novel inhibitors on SARS CoV 2 main protease receptor (PDB ID: 6XBH). The docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with designed novel inhibitors with Index numbers 46d, 741a, and 847b are presented in Figs. 8, 9 and 10, while Tables 9, 10 and 11 present interaction types with surrounding amino acids of SARS CoV 2 Main Protease (PDB ID: 6XBH) with designed novel inhibitor with Index numbers 46d, 741a and 847b.
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 46d (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 46d b 3D view of inhibitor with Index number 46d with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 46d with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 741a (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 741a; b 3D view of inhibitor with Index number 741a with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 741a with surrounding amino acids of 6XBH
Docked poses of SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor with Index number 847b (stick figure) a 3D view of docked pose of SARS CoV 2 Main Protease (PDB ID: 6XBH) with inhibitor with Index number 847b; b 3D view of inhibitor with Index number 847b with surrounding amino acids of 6XBH; c 2D view of interaction type of inhibitor with Index number 847b with surrounding amino acids of 6XBH
Discussion
The inhibitor with Index number 331 (IUPAC name 3,3'-((4-((4-hydroxy-2-oxo-2H-chromen-3-yl) (2-hydroxy-4-oxo-4H-chromen-3-yl)methyl)phenyl)methylene)bis(4-hydroxy-2H-chromen-2-one)), has the highest ICM score in magnitude, given as -48.38 kCal/mol. This is not a surprise as it has nine (9) hydrogen bonds, comprising of seven (7) conventional H-bonds involving Thr26, Gly143, Ser144, Cys145, Hie164, Thr25, and two (2) Carbon H-bonds involving Thr25 and Thr26. Further enquiry indicates the presence of an amide pi stacked interactions with Thr24 and Thr25, π-lone pair interaction with Thr24, π-pi stacked interaction with Hie41 and π-alkyl interaction involving Met49 and Cys145. Another noticeable point is the bond length of two of the conventional H-bonds involving Thr26 and Hie164 with bond length 1.68 and 1.94A, respectively, thus impacting positively on its activity. This is clearly shown in Fig. 2 and Table 2.
The docked structure presented in Fig. 3 and Table 3 showing interaction type of is 3-(4-(2-((tert-butoxycarbonyl) amino)-4-((3-((tert-butoxycarbonyl) amino)-2-hydroxy-4-phenylbutyl) amino)-3-hydroxybutyl) phenoxy) propanoic acid (inhibitor with Index number 441) with SARS CoV 2 main protease shows a negative free energy of binding (-29.01 kCal/mol) implying that binding is feasible as most of the interaction energies are of H-bond type with amino acids (Thr24, Thr25, Thr26, Gly143, Gln189, Hie164, Cys22) resulting in an overall negative value.
The result could be partly explained by the fact that the inhibitor has a strong hydrogen bond interaction with the amino acids of the binding pocket of the SARS CoV 2 main protease which is evidenced by the high hydrogen bond energy value of − 11.45 kCal/mol making it the highest of all the docked inhibitors. Other noticeable interactions with the receptor include carbon–hydrogen interaction with Thr26 and Thr25, π-sulfur interaction with Met49, amide-pi Stacked interaction with Leu141 and Asn142, π-alkyl interaction with Hie41. Zhijian Xu et al. in their work used both MM/GBSA and SIE methods and they voted for nelfinavir, with the binding free energy of − 24.69 ± 0.52 kCal/mol and − 9.42 ± 0.04 kCal/mol, respectively, to be a potential inhibitor against 2019- nCov Mpro (Xu et al. 2020). The inhibiting capacity of their proposed drug is not comparable to that obtained with the compounds with index numbers 441 and 741, making it a better drug candidate than nelfinavir.
The result for compound with index numbers 441 is shown in Fig. 4, the binding energy is reported in Additional file 1: Table S2 to be − 15.67 kCal/mol and the interaction type result is as shown in Table 4. The docked result shows that the inhibitor has seven hydrogen bond interactions with five amino acids (Thr26, Cys44, Gln189, Thr25, and Met49). The binding energy of inhibitor with Index number 741 is determined to be − 47.88 kCal/mol. This makes it the 2nd most active chemical agent with the ability to inhibit SARS CoV 2 main protease (Additional file 1: Table S2). The docked result owes its binding affinity to the presence of seven H-bond with the amino acids which include Thr26, Glu166, Hie164, and Met165 (Fig. 5). Other interactions such as Π-sulfur-type with Cys145, Met165, π-pi stacked with Hie41 and π-alkyl with Met49 are also observed (Table 5).
Based on hydrogen bond energy ranking, it occupies the third position, and it has hydrogen bond energy of 9.57 kCal/mol. Other important interactions such as π-alkyl, π-sulfur interactions are also reported. This is far better than all the compounds obtained by Motiwale and colleagues (Motiwale et al. 2020). They applied molecular docking approach in conjugation with molecular dynamics (MD) simulations to find out potential inhibitors against Mpro of SARS CoV-2 from previously reported SARS-3CL protease inhibitors.
They used a total of 61, previously known inhibitors. According to the molecular docking results, it was found that the binding energy of co-crystallized ligand, JFM (N-(2-phenylethyl)- methanesulfonamide) was found to be − 5.1 kCal/mol while the binding energy of all the 61 inhibitors lies between − 6.2 and − 8.8 kCal/mol. Where, 4-{[(4Z)-1-(3-chlorophenyl)-5-oxo-3-phenyl-4,5-dihydro-1H-pyrazol-4-ylidene]-methyl}benzoic acid, and 4-(4-methoxyphenyl)-6-oxo-2-[(2-phenylethyl)sulfanyl]-1,6-dihydropyrimidine-5-carbonitrile were reported to have minimum and maximum binding energy, respectively. Compounds having a binding energy of − 8.5 kCal/mol or less were considered better agents for the Mpro. Using this criteria, six molecules namely 4-{[(4Z)-1-(3-chlorophenyl)-5-oxo-3-phenyl-4,5-dihydro-1H-pyrazol-4-ylidene]methyl}benzoic acid, 5-amino-1-[2-(1-benzothiophen-2-yl)-2-oxoethyl] -2,3-dihydro-1H-indole-2,3-dione, N-(4-{[(4Z)-5-oxo-1,3-diphenyl-4,5-dihydro-1H-pyrazol-4-ylidene] methyl} phenyl) acetamide, (4Z)-4-{[4-(dimethylamino) phenyl]methylidene}-1,3-diphenyl-4,5-dihydro-1H-pyrazol-5-one, 4-{[(4Z)-5-oxo-1,3-diphenyl-4,5-dihydro-1H-pyrazol-4-ylidene]methyl}benzoic acid, and 4-{[(4Z)-1-(4-chlorophenyl)-5-oxo-3-phenyl-4,5-dihydro-1H-pyrazol-4-ylidene]methyl}benzoic acid were selected as potential drug candidate (Motiwale et al. 2020).
SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor Index number 819
Benzyl (1-((3-hydroxy-5-((2-hydroxy-2,3,3a,7a-tetrahydro-1H-inden-1-yl) amino)-4-((4-methoxybenzyl) amino) -5-oxo-1-phenylpentan-2-yl) amino)-3,3-dimethyl-1-oxobutan-2-yl) carbamate (inhibitor Index number 819) binds at the SARS CoV 2 Main Protease (PDB ID: 6XBH) binding site and forms eleven (11) H-bonds with a critical amino acids residue in the binding domain of 6XBH, five (5) of which are Conventional H-Bonds involving Cys44, Glu166, Hie164, Gly143, Glu166 and six (6) carbon-hydrogen bonds involving Met165, Hie164, Met49, Asp187 and Met49 which are clearly shown in Fig. 6 and in Table 6. The ICM score for the best possible interaction pose was given as − 47.52 kCal/mol, and that makes it the third most active compound reported. However, there are two notable unfavorable interactions reported. These are unfavorable donor–donor and unfavorable acceptor–acceptor interactions. Other interactions include Π-alkyl interaction with Met49, Met165, Π-Pi stacked interaction with Hie41 and alkyl-type interaction with Cys44, Met49, Pro52.
SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor index number 847
As presented in Additional file 1: Table S2, the binding energy of this interaction is reported to be − 39.89 kCal/mol. This is the inhibitor with the highest number of hydrogen bond making it one of the best chemical agents with the ability to inhibit SARS CoV 2 main protease. The docked result owes its binding affinity to the presence of thirteen (13) H-bond with the amino acids Ser144, Cys145, Glu166, Gln189, His163, Hie41, Hie164, Gly143, Met165, Asn142, Leu141 as shown in Fig. 7 and confirmed in detail in Table 7. The inter-atomic distances for the H-bond are 2.514, 2.57, 1.866, 1.574, 2.402, 2.649, 2.2, 3.069, 2.676, 2.722, and 2.247 Å, respectively.
Other interactions such as π-anion interaction with Glu166 and π-alkyl interaction with Hie41, Met165 also contributed to the high affinity of the inhibitor in the binding site by stabilizing its structure to conform to the surface of the polar amino acids. From the virtual screening results by Khan and colleague, two drug molecules were selected for each drug target protein [Paritaprevir (ΔG = − 9.8 kCal/mol) &Raltegravir (ΔG = − 7.8 kCal/mol) for 3CLpro and Dolutegravir (ΔG = − 9.4 kCal/mol) and Bictegravir (ΔG = − 8.4 kCal/mol) for 2'-OMTase]. From their extensive computational analysis, they proposed Raltegravir, Paritaprevir, Bictegravir and Dolutegravir as excellent lead candidates for these crucial proteins and they could become potential therapeutic drugs against 2019-nCoV (Khan et al. 2020). This result cannot be compared with our proposed drug that has a binding free energy of − 39.89 kCal/mol.
SARS CoV 2 main protease (PDB ID: 6XBH) with inhibitor index number 741a
2-amino-2-hydroxy-1-phenylethyl (2-((4-(benzylamino)-5-((1-(benzylamino)-1-oxobutan-2-yl) amino) -3-hydroxy-5-oxo-1-phenylpentan-2-yl)amino)-2-oxo-1-phenylethyl) carbamate binds firmly at the target site of 6XBH with seven Conventional H-Bonds (Thr26, Gly143, Gln189, Glu166, Thr24) and four C-H interaction with Thr26, Asn142, Hie164. The ICM score for the best interaction pose is reported in Table 8 as − 45.33 kcal/mol.
This reported result for the interaction of 2-amino-2-hydroxy-1-phenylethyl (2-((4-(benzylamino)-5-((1-(benzylamino)-1-oxobutan-2-yl)amino)-3-hydroxy-5-oxo-1-phenylpentan-2-yl)amino)-2-oxo-1-phenylethyl) carbamate in the binding site of 6XBH in Table 9 is attributed to the large number of π-interactions such as π-pi interaction with Hie41, π-alkyl interaction with Met49, Met165, and Leu167, amide-pi stacked interaction with Leu145, Asn142, Gly143 and finally π-sulfur interaction with (Met49 and Met165). However, there is also an unfavorable donor–donor interaction with 6XBH which was the reason further studies was not carried out on it (Fig. 8).
The docked structure presented in Fig. 9 and Table 10 showing interaction type of SARS CoV 2 Main Protease (PDB ID: 6XBH) with 2-(2-(5-amino-2-((((3-aminobenzyl) oxy) carbonyl) amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid (Index number 847b) shows a binding energy of − 41.32 kCal/mol implying that binding is feasible as most of the interaction energies are of H-bond type with amino acids (Thr26, Gly143, Ser144, Cys145, Glu166, Gln189, Hie164, Met49, Thr26, Thr25, Thr190, Asn142, Met165) resulting in an overall negative value. The result could be partly explained by the fact that the inhibitor has nineteen (19) hydrogen bond interaction with the amino acids of the binding pocket of the SARS CoV 2 main protease which is evidenced by the high hydrogen bond energy value of − 16.31 kCal/mol making it the highest of all the newly designed inhibitors.
Other noticeable interactions with the receptor include π-alkyl interaction mediated through Cys145. The inhibitor benzyl (5-amino-1-((4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxy-1-phenylbutan-2-yl)amino)-1,5-dioxopentan-2-yl)carbamate (Index no. 847) from which it was designed has binding score energy of − 39.89 kCal/mol and H-bond energy of − 10.27 kCal/mol as against binding score energy and H-bond energy of − 41.32 and − 16.31 kCal/mol, respectively, for the novel inhibitor (Table 11). Komatsu et al. in their work, show the binding pose of main protease system of SARS CoV 2 with darunavir, the ligand interacted with Ser46, Met49, Glu166, Val186, Gln189, and Thr190, ritonavir interacted with Cys44, Cys145, Met165, Asp187, Arg188, and Gln189, indinavir interacted with His41, Gly143, Glu166, and Gln189 (Komatsu et al. 2020).
None of these proposed drugs have as much interactions as 2-(2-(5-amino-2-((((3-aminobenzyl)oxy) carbonyl)amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid (the novel inhibitor). This improvement resulted from the primary amine group attached to meta position of first benzene ring and the carboxyl group attached to the ortho position of the second benzene ring (Fig. 10). This result makes 2-(2-(5-amino-2-((((3-aminobenzyl)oxy) carbonyl) amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid a better drug candidate against SARS CoV-2 main protease in comparison with the co-crystallized inhibitor or any of the 1000 inhibitors.

Molecular dynamics
Figure 11 shows the 2D interaction of REF-IN with the main protease of SARS CoV2 before and after molecular dynamics study, while Figs. 12 shows the RMSD plot. Figures 13 and 14 shows the shows the 2D interaction of inhibitor with Index number 847 with the main protease of SARS CoV 2 before and after molecular dynamics study, a plot of internal energy with time, a plot of potential energy with time, and a plot of enthalpy change with time, respectively. Figures 15 and 16 show the 2D interaction of inhibitor 847b with the main protease of SARS CoV 2 before and after molecular dynamics study, and a plot of internal energy against time.
Reference inhibitor
Interaction of benzyl (5-amino-1-((4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxy-1-phenylbutan-2-yl) amino)-1,5-dioxopentan-2-yl) carbamate (inhibitor Index number 847) before and after molecular dynamics study
Figure 13 is the 2d interaction of SARS CoV 2 main protease with benzyl (5-amino-1-((4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxy-1-phenylbutan-2-yl)amino)-1,5-dioxopentan-2-yl)carbamate (inhibitor Index number 847) before and after molecular dynamics study placed side by side. It can be seen that all the interactions remained intact after the dynamics study. This is further proven in Fig. 14 by the constancy of potential energy over time. The internal energy decreased consistently until 600 picoseconds when it stabilizes at − 264,500 kCal/mol.
Interaction of 2-(2-(5-amino-2-((((3-aminobenzyl)oxy) carbonyl) amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid (index number 847b) before and after molecular dynamics study.
Figures 15 and 16 show the 2d interaction of SARS CoV 2 main protease with 2-(2-(5-amino-2-((((3-aminobenzyl)oxy) carbonyl)amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl)benzoic acid (the novel inhibitor) before and after molecular dynamics study and the plot of internal energy versus time. As shown in Fig. 16, the internal energy decreases consistently until it stabilizes between 575 and 600 picoseconds. It also shows that the interaction is a spontaneous one in which energy in form of heat is lost, the enthalpy change reduces gradually over time to achieve stability at 600 picoseconds.
Conclusions
The molecular docking results shown in the figures confirm that the hydrophobic and hydrogen bonding interactions with these targets have pivotal contributions to the binding structures and binding free energies, even though the van der Waals and π-interactions contributed to the stabilization of the binding structures.
The molecular docking result also shows that, inhibitors with Index numbers 331, 741, 819, 441, 847, and 46 with ICM score of − 48.38 kCal/mol, − 47.88 kCal/mol, − 47.52 kCal/mol, 29.01 kCal/mol, 39.89 kCal/mol, and − 15.67 kCal/mol, respectively, best inhibit SARS CoV 2 main protease of the compounds within our data set. These compounds were further utilized in designing new potent inhibitor compounds by attaching potent fragments to the compounds. Most of the newly designed compounds were reported to be more active than the parent structure. This includes compounds with index number 741a, 847b, and 741d with a binding affinity of − 45.33 kCal/mol, − 41.32 kCal/mol and − 40.12 kCal/mol, respectively. However, compounds with index numbers 741a and 741b and 46d were not considered to be our potential drug candidate because of the presence of unfavorable interactions they formed with SARS CoV2 main protease. The fragments responsible for their affinities were primarily carboxylic group and primary amine group. At the end of the study, we were able to computationally design a potent novel compounds that can be used to inhibit SARS CoV 2 main protease. The novel drug is 2-(2-(5-amino-2-((((3-aminobenzyl)oxy) carbonyl) amino)-5-oxopentanamido)-4-(2-(tert-butyl)-4-oxo-4-(pentan-3-ylamino) butanamido)-3-hydroxybutyl) benzoic acid with binding score energy and H-bond energy of − 41.32 and − 16.31 kCal/mol, respectively.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Abbreviations
- AMBER:
-
Assisted model building with energy refinement
- Å:
-
Armstrong
- BFGS:
-
Broyden–Fletcher–Goldfarb–Shanno (BFGS) algorithm
- CADD:
-
Computer-aided drug design
- CHARMM:
-
Chemistry at HARvard macromolecular mechanics
- CG:
-
Conjugate gradient
- DNA:
-
Deoxyribonucleic acid
- GROMOS:
-
GROningen MOlecular Simulation
- GUI:
-
Graphic user interphase
- HCV:
-
Hepatitis C virus
- HIV:
-
Human immunodeficiency virus
- ICM:
-
Internal coordinate mechanics
- kCal/mol:
-
Kilo Calorie per mole
- LBDD:
-
Ligand-based drug design
- L-BFGS:
-
Limited memory Broyden–Fletcher–Goldfarb–Shanno (BFGS) algorithm
- LGA:
-
Lamarckian genetic algorithm
- MD:
-
Molecular dynamics
- MM-GBSA:
-
Molecular mechanics/generalized born surface area
- MM:
-
Molecular mechanics
- Mpro:
-
Main protease
- MT-DTI:
-
Molecule transformer–drug target interaction
- MTTK:
-
Martyna–Tuckerman–TobiasKlein
- NAMD:
-
NAnoscale molecular dynamics
- nCoV:
-
Novel corona virus
- NMR:
-
Nuclear magnetic resonance
- NVE:
-
Microcanonical ensemble (a system is isolated from changes in moles (N), volume (V), and energy (E))
- NVT:
-
Canonical ensemble (amount of substance (N), volume (V) and temperature (T) are conserved)
- NS3:
-
Non-structural protein 3
- PBC:
-
Periodic boundary conditions
- PDB:
-
Protein data bank
- PM6:
-
Parameterization method 6
- PME:
-
Particle mesh Ewald
- QSAR:
-
Quantitative structure–activity relationship
- RdRp:
-
RNA-dependent RNA polymerase
- RMSD:
-
Root mean square deviation
- RNA:
-
Ribonucleic acid
- SARS CoV 2:
-
Severe acute respiratory syndrome coronavirus 2
- SBDD:
-
Structure-based drug design
- SD:
-
Steepest descent
- VMD:
-
Visual molecular dynamics
- 0D, 1D, 2D, 3D:
-
Zero- , one- , two- and three-dimensional descriptor, respectively
- 2’-O-MTase:
-
2’-O-ribose methyltransferase
- 3 CL pro:
-
3 C-like proteinase
- ∆G:
-
Free energy of binding
References
Abagyan R, Totrov M, Kuznetsov D (1994) ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15(5):488–506
Abd El-Aal M, Mogharbel RT, Ibrahim A, Almutlaq N, Zoromba MS, Al-Hossainy AF, Ibrahim SM (2022) Synthesis, characterization, and photosensitizer applications for dye-based on ZrO2-acriflavine nanocomposite thin film [ZrO2+ ACF] C. J Mol Struct 1250:131827
Adeniji SE, Uba S, Uzairu A (2020) Multi-linear regression model, molecular binding interactions and ligand-based design of some prominent compounds against Mycobacterium tuberculosis. Network Model Anal Health Inform Bioinform 9(1):1–18
Al-Hossainy AA, Ibrahim A, Mogharbel RT, Ibrahim SM (2021) Synthesis of novel keto-bromothymol blue in different media using oxidation–reduction reactions: combined experimental and DFT-TDDFT computational studies. Chem Pap 75(7):3103–3118
Arthur DE, Ejeh S, Uzairu A (2020) Quantitative structure-activity relationship (QSAR) and design of novel ligands that demonstrate high potency and target selectivity as protein tyrosine phosphatase 1B (PTP 1B) inhibitors as an effective strategy used to model anti-diabetic agents. J Recept Signal Transd 40(6):501–520
Arthur DE, Uzairu A (2019) Molecular docking studies on the interaction of NCI anticancer analogues with human Phosphatidylinositol 4, 5-bisphosphate 3-kinase catalytic subunit. J King Saud Univ-Sci 31(4):1151–1166
Arthur DE, Uzairu A, Mamza P, Abechi SE, Shallangwa GA (2018) Structure-based optimization of tyrosine kinase inhibitors: a molecular docking study. Network Model Anal Health Inform Bioinform 7(1):1–18
El Azab I, Thabet HK, Almotairi SA, Saleh M, Mogharbel R, Mahmoud S, Abdel-Aziz M et al (2021) Synthesis of a novel coumarin heterocyclic derivative and fabrication of hybrid nanocomposite thin film with CoOFe2O4 for optoelectronic applications. J Mol Struct 1241:130640
Baig MS, Alagumuthu M, Rajpoot S, Saqib U (2020) Identification of a potential peptide inhibitor of SARS-CoV-2 targeting its entry into the host cells. Drugs R&d 20(3):161–169
Berendsen HJ, Postma J, V, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690
Bikadi Z, Hazai E (2009) Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 1(1):1–16
Biovia DS (2017) Discovery studio visualizer. San Diego 936
DiMasi JA, Hansen RW, Grabowski HG (2003) The price of innovation: new estimates of drug development costs. J Health Econ 22(2):151–185
Du Y, Tu L, Zhu P, Mu M, Wang R, Yang P, Hu P et al (2020) Clinical features of 85 fatal cases of COVID-19 from Wuhan. A retrospective observational study. Am J Respirat Crit Care Med 201(11):1372–1379
Environment MO (2014). Molecular Operating Environment (MOE), 2014.09. In: Chemical Computing Group Inc
Gagic Z, Ruzic D, Djokovic N, Djikic T, Nikolic K (2020) In silico methods for design of kinase inhibitors as anticancer drugs. Front Chem 7:873
Gallicchio E, Lapelosa M, Levy RM (2010) Binding energy distribution analysis method (BEDAM) for estimation of Protein−Ligand binding affinities. J Chem Theory Comput 6(9):2961–2977
De Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Memish ZA et al (2013) Commentary: middle east respiratory syndrome coronavirus (mers-cov): announcement of the coronavirus study group. J Virol 87(14):7790–7792
Haspel N, Zheng J, Aleman C, Zanuy D, Nussinov R (2017) A protocol for the design of protein and peptide nanostructure self-assemblies exploiting synthetic amino acids. In: Computational protein design. Springer, pp 323–352
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Bioinform 65(3):712–725
Hosseini FS, Amanlou M (2020) Anti-HCV and anti-malaria agent, potential candidates to repurpose for coronavirus infection: Virtual screening, molecular docking, and molecular dynamics simulation study. Life Sci 258:118205
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Gu X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Ibrahim SM, Bourezgui A, Al-Hossainy AF (2020) Novel synthesis, DFT and investigation of the optical and electrical properties of carboxymethyl cellulose/thiobarbituric acid/copper oxide [CMC+ TBA/CuO] C nanocomposite film. J Polym Res 27(9):1–18
Khan RJ, Jha RK, Amera GM, Jain M, Singh E, Pathak A, Singh AK, et al (2020) Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J Biomol Struct Dyn 1–14
Komatsu TS, Okimoto N, Koyama YM, Hirano Y, Morimoto G, Ohno Y, Taiji M (2020) Drug binding dynamics of the dimeric SARS-CoV-2 main protease, determined by molecular dynamics simulation. Sci Rep 10(1):1–11
Lau SK, Woo PC, Li KS, Huang Y, Tsoi H-W, Wong BH, Yuen K-Y et al (2005) Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci 102(39):14040–14045
Mohamed NS, Ahmed MM, Yahia A, Ibrahim SM, Al-Hossainy AF (2022) Development of azithromycin–Pd mono nanocomposite: Synthesis, physicochemical, characterization and TD-DFT calculations. J Mol Struct 1263:133126
Motiwale M, Yadav NS, Kumar S, Kushwaha T, Choudhir G, Sharma S, Singour PK (2020) Finding potent inhibitors for COVID-19 main protease (Mpro): an in silico approach using SARS-CoV-3CL protease inhibitors for combating CORONA. J Biomol Struct Dyn 1–12
Phillips JC, Hardy DJ, Maia JD, Stone JE, Ribeiro JV, Bernardi RC, Jiang W et al (2020) Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys 153(4):044130
Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, Corman VM et al (2013) Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 13(10):859–866
Sacco MD, Ma C, Lagarias P, Gao A, Townsend JA, Meng X, Kitamura N, et al (2020) Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci Adv 6(50):eabe0751
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234
Stockwell BR (2000) Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet 1(2):116–125
Viswanadhan VN, Ghose AK, Revankar GR, Robins RK (1989) Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J Chem Inform Comput Sci 29(3):163–172
Wavefunction I (2013) Spartan'14, version 1.1. 2. In: California, USA Irvine
Xu Z, Peng C, Shi Y, Zhu Z, Mu K, Wang X, Zhu W (2020) Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation
Zwawi M, Attar A, Al-Hossainy A, Abdel-Aziz M, Zoromba MS (2021) Polypyrrole/functionalized multi-walled carbon nanotube composite for optoelectronic device application. Chem Pap 75(12):6575–6589
Acknowledgements
We would like to thank the Head of Department of Pure and Applied Chemistry, UNIMAID as well as the Head of Department BAZE University for their continuous support throughout the period of this research
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
DEA, MS and AOA conceived the work, DEA, MS and BOE carried out the research, while DEA, and BOE wrote and edited the paper, all authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
No competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplementary table 1: Name, target source, article doi, Authors and Zinc ID of the complete dataset. Supplementary table 2. Molecular docking result of reference inhibitor and complete dataset on COVID 19 main protease receptor (PDB ID: 6XBH). Supplementary table 3: Structure and IUPAC Name of Designed Novel Inhibitors.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arthur, D.E., Elegbe, B.O., Aroh, A.O. et al. Computational drug design of novel COVID-19 inhibitor. Bull Natl Res Cent 46, 210 (2022). https://doi.org/10.1186/s42269-022-00892-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-022-00892-z