Abstract
Background
Carbapenem antibiotics consider the primary treatment choice for serious Pseudomonas aeruginosa infection. Hence, the evolution of carbapenem resistance mediated by acquiring genes encoding class b enzymes is of global concern. The purpose of this article research is to explore the prevalence, drug resistance profiles, and metallo-β-lactamases (MβLs) production in extensively drug-resistant carbapenem-resistant P. aeruginosa (XDR-CRPA).
Methods
P. aeruginosa isolates were collected and identified according to conventional methods. Antibiotic resistance patterns were determined by single disk diffusion. Minimum inhibitory concentrations (MICs) of (imipenem, meropenem, ceftazidime, piperacillin/tazobactam, levofloxacin, and gentamicin) were determined for CRPA. A subset of the isolates collection consisting of the XDR-CRPA with the highest MICs to imipenem and meropenem were selected for the phenotypic screening of carbapenemases and MβLs production capability using the modified carbapenem inactivation (mCIM) and imipenem-EDTA combined disk (MβL-CD) methods, respectively. Then, molecular analysis, including identification by the specific primer of 16S rRNA and detection of MβL genes using polymerase chain reaction (PCR) was performed to the XDR selected isolates.
Results
Among 100 P. aeruginosa isolated throughout this period, 59% exhibited reduced susceptibility rates to carbapenems. A total of 20.3% and 57% of CRPA isolates were MDR and XDR, respectively. MIC values of the CRPA revealed that these isolates exhibited high MIC50 and MIC90 to the six selected antibiotics. The findings of the (mCIM) assay displayed identical concordance results with the MβL-CD. Molecular investigation technique assured that 10 (90.9%) and 2 (18.1%) of the 11 XDR selected isolates are positive for blaNDM-1 and blaVIM-1 genes, respectively. Polymyxin B and colistin followed by aztreonam were the most effective antibiotics used for curing infections caused by XDR Pseudomonas aeruginosa.
Conclusion
The prevalence of high XDR-CRPA in our study is a critical problem. Our present study found that the blaNDM-1 was present at a significant frequency among the selected XDR isolates, highlighting the need for establishing strict antimicrobial policies to avoid the prompt spread of these isolates.
Similar content being viewed by others
Introduction
Pseudomonas aeruginosa is an opportunistic gram-negative rod, which regarded as a significant reason for an ever-widening array of various life-threatening infections (Esmaeili et al. 2019). As a matter of fact, this bacterium is the main causing of burn infections and ventilator-associated pneumonia (VAP). Moreover, this microorganism can potentially initiate infections in the urinary tract, lung, soft tissue, and abscess (Oliver et al. 2008).
Carbapenems are a powerful member of the beta-lactam class with a broader antibacterial spectrum, knowing in treating serious and critical infections. However, the development of reduced susceptibility to this life-saving antimicrobial has been increasingly reported in Pseudomonas aeruginosa (Feng et al. 2017). Different mechanisms lead to the evolution of carbapenem-resistant P. aeruginosa especially through the acquiring of resistant genes encoding carbapenem-hydrolyzing enzymes (Shaaban et al. 2017). Among them, the metallo-beta-lactamases (MβLs) (e.g., VIMs, IMPs, NDMS) appeared to be the utmost importance enzymes for P. aeruginosa (Patel and Bonomo 2013). Most genes encoding MβLs are either plasmid- or chromosome-mediated, located as horizontally transferable cassettes typically cluster with other drug resistance determinants. These reasons explain why the spread of P. aeruginosa with MβL activity can result in the appearance of the pan-resistant phenotype, leaving limited number or even non-therapeutic options available (Dogonchi et al. 2018).
New Delhi metallo-beta-lactamase-1 (NDM-1), the recently emerged MβL, can deactivate nearly to all beta-lactams including carbapenems with excluding of monobactam. The first discovery of bacterial isolate carrying this novel enzyme was from more than 10 years in India (Yong et al. 2009). Until now, NDM-1-producing-bacteria have been raised over 40 countries that pose a tremendous public health risk (Jamal et al. 2016). The first detection of NDM-1variant in Egypt was in Acinetobacter baumannii isolate (Kaase et al. 2011).
Another MβL family includes Verona integron metallo beta-lactamase (VIM) coding gene determinant has likewise been identified primarily among carbapenem-resistant P. aeruginosa from Southern Europe. The blaVIM-1 gene was first detected in Italy, to date, the VIM group has included 37 variants (Levy Hara et al. 2013; Walsh 2013). In this study, antimicrobial susceptibilities to different anti-pseudomonal antibiotics were compared in P. aeruginosa isolates in Egypt, with a specific focus on the categorization of the concerned isolates to MDR and XDR phenotypes. Secondly, quantitative MIC determination of the main antibiotics’ groups against carbapenem-resistant isolates was investigated. Finally, phenotypic and genotypic techniques were used for assessing the emerging of metallo-β-lactamase-producing isolates in a collection of extensive drug-resistant P. aeruginosa.
Materials and methods
Bacterial isolates
Pseudomonas aeruginosa isolates were collected during the period of May to September 2017 from the microbiological laboratory in (Kasr El Aini Hospital, Cairo University, Giza, Egypt) and (El Borg Laboratory, Mohandseen, Giza, Egypt).
Conventional methods for identification
Isolates were identified by standard microbiological and biochemical methods based on gram staining technique, colonial morphology (5% citrated horse blood agar and MacConkey’s agar, Oxoid, ltd., Basin Stoke, Hants, England), motility determination using the agar stabbing method, indole, fluorescein and pyocyanin pigment production, catalase and oxidase reactions, oxidative-fermentative tests, citrate utilizing, nitrate reduction, acetamide hydrolysis and growth at 42 °C (Engelkirk and Duben-Engelkirk 2008; Mahon et al. 2014).
Antimicrobial susceptibility profiling
Antimicrobial resistance was applied by disk diffusion method using the Kirby Bauer technique (Bauer et al. 1966) and as per the recommendations and interpretations of a CLSI document (CLSI 2009; CLSI 2018) using the following antibiotic disks (Oxoid ltd., Basin Stoke, Hants, England): imipenem (IPM 10 μg), piperacillin-tazobactam (TZP 100 μg/10 μg), meropenem (MEM 10 μg), piperacillin (PRL 100 μg), ceftazidime (CAZ 30 μg), aztreonam (ATM 30 μg), cefepime (FEP 30 μg), gentamicin (CN 10 μg), levofloxacin (LEV 5 μg), amikacin (AK 30 μg), and ciprofloxacin (CIP 5 μg), ATCC strain 27853 was examined routinely to monitor the test performance.
For the susceptibility testing to Polymyxin B, and colistin antibiotics (Biotika and Waki pharma, Egypt, respectively) against P. aeruginosa isolates, minimum inhibitory concentrations (MICS) were detected using the broth microdilution technique as described by Andrew and CLSI guidelines (Andrews 2001; CLSI 2012; CLSI 2018). After an overnight incubation at 37 °C, the viability of cells was detected by adding 20 μl of triphenyl tetrazolium chloride dye solution (5 mg/ml) (TTC) to each well (Cat. No. 1500 A, research lab fine chemical industries, India) (Manzoor et al. 2016). P. aeruginosa isolates were subsequently categorized according to the new standardized international document (Magiorakos et al. 2012) to MDR and XDR isolates.
MIC distributions of selected antibiotics against carbapenem-resistant P. aeruginosa
The MICs were detected to six selected antibiotics related to five main groups. The following antibiotics were obtained from Egyptian pharmaceutical companies: imipenem from Soficopharm; meropenem triydrate from Astrazeneca; piperacillin/tazobactam from Hikma pharmaceuticals; ceftazidime pentahydrate from Smith Kline Beecham; gentamicin sulfate from Memphis and levofloxacin hemihydrates from Sanofi-Aventis. The values of MIC50 and MIC90 were calculated for all the antibiotics tested.
Phenotypic tests
Phenotypic detection for carbapenemase production
XDR carbapenem-resistant P. aeruginosa (XDR-CRPA) with the highest MIC to imipenem and meropenem antibiotics were selected to survey the occurrence of carbapenemase enzymes using modified carbapenem inactivation method (mCIM) according to CLSI M100-S28 guidelines (CLSI 2018). The result was considered to be positive for the inhibition zone diameter of 6–15 mm, negative for ≥19 mm.
Phenotypic screening of metallo-β-lactamase (MβLs) activity
All the carbapenemase positive P. aeruginosa were checked for the production of MβLs by imipenem-EDTA combined disk (MβL-CD) method as described previously (Pitout et al. 2005). More than or equal to7 mm zone enhancement in IPM-EDTA disk compared to the aligned IPM disk alone judged to indicate positivity for MBL.
DNA extraction
The genomic DNA was prepared by the boiling lysis method (Moore et al., 2004). A few colonies of an overnight culture were dispensed in 250 μl of Tris EDTA 1× solution buffer, the suspension was heated in a thermal block (HB-200, Human lab instruments, Korea) at 100 °C for 8 min, then placed at −20 °C for 5 min. Following this, the supernatant was centrifuged at 15,000 rpm at 4 °C for 60 s to remove cellular debris. An aliquot of the supernatant was used as an origin for DNA template.
PCR methods
PCR amplification was performed in a thermocycler (PTC-100, MJ Research, Waltham, MA, USA). The amplification was in a total volume of a 25 μl containing 12.5 μl of RedTaq PCR Master Mix (3 mM MgCl2, 0.4 mM of each dNTP, 0.06 U/μl of Taq DNA polymerase), (Sigma-Aldrich, California, USA), 1 μl of each forward and reverse primer (10 μM), (Microsynth, Balgach, Switzerland), 3 μl of whole-cell bacterial lysate, and 7.5 μl nuclease-free water. The PCR products were separated by gel electrophoresis (DYCP-31DN, Beijing Liuyi instrument, Beijing, China), with 1.6% agarose gel (Fisher Bio Reagents, USA), then viewed under an ultraviolet (UV) transilluminator (LMS-20E UVP, Upland, California, USA). Molecular size marker (100 bp) (Direct load, Sigma Aldrich, California, USA).
Molecular identification
The selected XDR-CRPA isolates were identified by the molecular approach through amplification of 16 s ribosomal RNA gene with a specific primer described by Spilker et al. 2004. As a reference strain, P. aeruginosa PAO1 was kindly provided by (Dr Mona Shaaban, Department of Microbiology, Faculty of Pharmacy, Mansoura University).
Molecular detection of the MβLs encoding genes
The selected XDR-CRPA isolates were screened for the presence of the MβLs encoding genes; blaVIM-1, blaNDM-1, and blaIMP by polymerase chain reaction (PCR) using specific primers for amplification each gene. The primers used were NDM-1, VIM-1, VIM-2, and IMP as described previously (Shaaban et al. 2017; Poirel et al. 2011) (Table 1).
Statistical analysis
Statistics were performed using Pearson’s chi-squared test to assess the significance of differences. A probability of less than 0.05 was considered statistically significant.
Results
Bacterial identification
Totally one hundred nonconsecutive P. aeruginosa isolates were collected during 4 months of study; Fig. 1 showed randomly selected Pseudomonas aeruginosa isolates on different culture media.
The colonial morphology of P. aeruginosa isolates on different culture media. a Colorless colonies of P. aeruginosa on MacConkey agar. b Blue pyocyanin pigment production on Pseudomonas aeruginosa (P) agar. c Green metallic sheen colonies of P. aeruginosa on citrated horse blood agar. d The fluorescence characteristics of P. aeruginosa on cetrimide agar under the short wavelength of ultraviolet light
Antimicrobial susceptibility profile
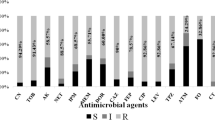
According to results from the antibiotic susceptibility test; resistance rates for β-lactam antibiotics were 74% (cefepime), 64% (piperacillin), (55%) piperacillin/tazobactam 53%, and (ceftazidime), while low resistance rates were observed to aztreonam (23%). For non-β-lactam antibiotics, the resistance rates were 70% (gentamicin), 67% (levofloxacin), 66% (amikacin), and 64% (ciprofloxacin). Among our batch of isolates, polymyxin b and colistin retained in vitro activity against all isolates (MIC range was 0.5–2 μg/ml). Overall, a significantly greater number of CRPA isolates were more resistant to the most of the antibiotics than carbapenem susceptible P. aeruginosa isolates (CSPA) (P < 0.05) (Table 2). Of all the isolates tested, we classified twenty-one (21%) isolates as (MDR) and forty-seven (47%) as (XDR). Among CRPA, the percentages of MDR-PA and XDR-PA were 20.33% and 79.66%, respectively. The XDR and MDR isolates were grouped into 23 resistance profiles, with profile 4 being predominant and accounting for 57.6% (34/59) of all carbapenem isolates. This profile showed resistance to 10 antibiotics (IPM, MEM, CIP, CN, CAZ, PRL, TZP, LEV, AK, FEP) (Table 3).
MIC distributions
Tables 4 and 5 showed the MIC distributions of six antimicrobial agents against the 59 CRPA isolates. For imipenem, MICs at which 90% of isolates inhibited were markedly higher (1024 μg/ml) whereas the MIC90 values for the meropenem antibiotic were only 2-fold lower than those of imipenem. Additionally, we noted that (42.3%) of isolates had an imipenem MIC equal to1024 μg/ml and (25.4%) had a meropenem MIC = 512 μg/ml. Furthermore, our results revealed that 2 (3.4%) were susceptible to meropenem with MIC range between (1 and 2 μg/ml), and three isolates (2.5%) had MIC = 4 μg/ml to imipenem and/or meropenem. In the CLSI guidelines, we could categorize these isolates as an intermediate resistance. The analysis of MIC data to the other antibiotics demonstrated that CRPA was non-susceptible to piperacillin/tazobactam with MIC90 superior or equal to 128/4 μg/ml. Furthermore, levofloxacin potency is almost negligible since all the isolates had MIC above the resistance breakpoints according to CLSI whereas the lowest MIC recorded was 16 μg/ml. The MIC range for gentamicin was 4–32 μg/ml in 22% of the isolates (n = 13), while,= 77.8% (n = 46) had MICs of ≥1024 μg/μl to this antibiotic. Similarly, the findings of MIC50 and MIC90 of ceftazidime were at the upper limit (both being ≥1024 μg/ml) of the breakpoint (Fig. 2). After MIC distribution results, we selected a set of 11 XDR isolates, which were had the highest MIC values to imipenem and meropenem antibiotics for further analysis.
Phenotypic tests
The mCIM showed the presence of carbapenemase activity in all the selected isolates. Moreover, it displayed an increase in inhibition zone (≥7 mm) around imipenem + EDTA disk, which lead to the interpretation that the isolates were MβL positive (Fig. 3).
Molecular microbiology results
Molecular identification based on PCR amplification of 16S rDNA
All the 11 selected XDR isolates have been confirmed by molecular identification as P. aeruginosa. They yielded fragment at the predicted size 956 bp (Fig. 4a) through amplification of 16 s rDNA region using species-specific primers for P. aeruginosa.
PCR analysis of P. aeruginosa isolates on agarose gel electrophoresis (a) PCR products for amplification-specific 16S rDNA region to P. aeruginosa. Lanes, 1-11: P. aeruginosa isolates; lane 12, PAO1 strain (positive control). b PCR products for amplification metallo-beta-lactamase gene encoding blaVIM-1. c PCR products for amplification metallo-beta-lactamase gene encoding blaNDM-1
Molecular detection of MβLs encoding genes
PCR assays showed 133 bp amplified band which was specific to blaNDM-1 gene in 10 (90.9%) out of 11 isolates and 920 bp targeting blaVIM-1 gene was present in 2 (18.1%) of isolates. One isolate co-harbored blaNDM-1 and blaVIM-1. In this work, blaIMP and blaVIM-2 allele were not detected in this study (Fig. 4b and c).
Discussion
The worldwide, worrying situation of reduced susceptibility to carbapenem in P. aeruginosa becomes noticed over the last decade, reveal a need for an exhaustive investigation. The incidence of resistance to carbapenems among P. aeruginosa was high (59%). This pace of resistance reflects a risk restricting the therapeutic options. It is reasonable to explain that this may be correlated with the extent use of carbapenems in recent years as well as a selective pressure from increased prescriptions of carbapenem. As anticipated, similar high rates of resistance have been recorded in Egypt (Gaballah et al. 2018; Abaza et al. 2017). The frequency of carbapenem resistance P. aeruginosa was also recognized in our Middle East region (Al-Agamy et al. 2011; Maroui et al. 2016) and worldwide (Castanheira et al. 2014).
It was important to note that CRPA was markedly more resistant than CSPA isolates for the most majority of antibiotics tested and the highest effective antibiotics against both groups were polymyxin B and colistin. Our observations concurred with other published reports (Kiddee et al. 2013; Dogonchi, et al. 2018).
XDR P. aeruginosa demonstrates a major point of interest worldwide. What is more, this bacterial phenotype could be the source of infections with an excessive mortality pace which leads to difficulty in the treatment (Derosa et al. 2019). Our data clearly indicate that the extremely high predominance of XDR among carbapenem-resistant isolates (79.6%).
The high rate of XDR-CRPA found in this study could be attributed to the incessant and uncontrolled consumption of antibiotics in Egypt; the well-known information is that antibiotics could be easily purchased without a doctor’s prescription in Egypt (what is known as patient’s self-medication), thus require increment monitoring and implementation of a strategy of antibiotic use. In contrast to our study, other works reported a low prevalence of XDR CRPA (Li et al. 2016; Shu et al. 2012).
Considering that 57.6% of the CRPA isolates in this report had an identical resistance profile, this led to the rise of a new drug-resistant pattern for carbapenem-resistant P. aeruginosa in our country.
The determination of MIC values affirmed the result identified by the single disk diffusion method. Having 33 isolates (55.9%) with a MIC of ≥256 μg/ml for one or more of the carbapenems tested is perturbing. Furthermore, high values of MIC for the rest of the antibiotics tested is a critical problem, increasing the widespread challenges in treatment, in this manner, it is indisputable that these drugs are not proper for serious infections via carbapenem-resistant P. aeruginosa. High MIC values for imipenem, meropenem, and ceftazidime antibiotics had been reported previously (Al-Agamy et al. 2016)
To enhance carbapenemase and metallo-β-lactamase detection, different phenotypic tests were suggested as the initial screening step prior using corroborative molecular investigations. Alongside the aforementioned, our results revealed that mCIM and MβL-CD tests are valuable and dependable with high accuracy and minimal cost; thus, it can be implemented in all labs and does not demand specific equipment. The results of carbapenemase and MβL obtained are according to other studies (Malkoçoğlu et al. 2017; Ismail et al. 2018).
According to the results, PCR asserted the presence of P. aeruginosa DNA in all the selected isolates identified conventionally. The confirmation of P. aeruginosa identification via molecular approach turned out to be increasingly imperative to diminish the issues of culture method such as a false negative culture result that may be owing to the sample overgrowth by other bacteria (Shahat et al. 2019). Of the acquired metallo-β-lactamase genes investigated, NDM-1 had been the most frequently detectable gene among the selected XDR-PA isolates, although the rare occurrence in Egypt. This result is of great concern and has shed light on the fact that NDM-1 producers are now alarmingly on the increase in our country. Few studies have reported the prevalence of NDM-1 in P. aeruginosa in Egypt (Zafer et al. (2014a); Shabaan et al. 2017). Furthermore, as our results revealed, two isolates carry the VIM-1 encoding gene in addition to one of them coexisted with the NDM-1 encoding gene. The co-harboring of two MβL genes in P. aeruginosa isolates has been reported in previous studies worldwide (Azimi et al. 2018; Li et al. 2016; Paul et al. 2016; and Rizek et al. 2014). None of the isolates demonstrated positive results for genes coding IMP-type MβL. This finding is in agreement with previous reports concluded the absence of this gene (Zafer et al. (2014b); Diab et al. 2013).
Antibiotic choice is particularly important in patients with menacing infections caused by CRPA. Stability to hydrolysis by MβL producers alongside its uncommon remedy in Egypt has settled on the aztreonam the subsequent option after polymyxins for treating gravely ill infected patients with multi-resistant P. aeruginosa.
Conclusion
In this context, our data may lead to three meaningful conclusions. First, the frequency of CRPA is observed. Besides, most of the carbapenem isolates are XDR. Thus, the presence of this high percent in such little sample size screened is of utmost concern. Second, all the eleven selected isolates were MβL producers. As regards, the resistance of MβL isolates to the most antibiotic groups, restricting the treatment alternatives for infections by these isolates. In this way, the early and accurate detection of MβLs with a straightforward and economical method ought to be executed for routine laboratory studies. Additionally, we propose regular control and continuous surveillance of highly resistant P. aeruginosa in medical institutions, as well as, the consciousness of high risk of self medications between Egyptians in the community settings will aid to forestall their dispersal and subsequently reduce the disease burden. Third, the most effective antibiotics that are evading XDR-CRPA are polymyxins. Nevertheless, excessive of these has caused to emerge polymyxins resistance. Thus, aztreonam could be used in treating of infections by XDR-CRPA isolates to reduce polymyxins usage.
Availability of data and materials
The datasets generated and/or analyzed during the current study are included in this published article
Abbreviations
- MBLs:
-
Metallo-β-lactamases
- XDR-CRPA:
-
Extensively drug-resistant carbapenem-resistant P. aeruginosa
- CSPA:
-
Carbapenem susceptible P. aeruginosa
- MICs:
-
Minimum inhibitory concentrations
- mCIM:
-
Modified carbapenem inactivation method
- MβL-CD:
-
Imipenem-EDTA combined disk method
- NDM-1:
-
New Delhi Metallo-beta-lactamase-1
- VIM:
-
Verona integron Metallo beta-lactamase
References
Abaza AF, Shazly SA, Selin HS, Aly GS (2017) Metallo-beta-lactamase producing Pseudomonas aeruginosa in a healthcare setting in Alexandria, Egypt. Pol J Microbiol 66:297–308
Al-Agamy MH, Jeannot K, El-Mahdy TS, Samaha HA, Shibl AM et al (2016) Diversity of molecular mechanisms conferring carbapenem resistance to Pseudomonas aeruginosa isolates from Saudi Arabia. Can J Infect Dis Med Microbiol. https://doi.org/10.1155/4379686
Al-Agamy MH, Shibl AM, Zaki SA, Tawfik AF (2011) Antimicrobial resistance pattern and prevalence of metallo-β-lactamases in Pseudomonas aeruginosa from Saudi Arabia. Afr J Microbiol Res 5(30):5528–5533
Andrews JM (2001) Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48:5–16
Azimi A, Peymani A, Pour PK (2018) Phenotypic and molecular detection of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from patients with burns in Tehran, Iran. Rev Soc Bras Med Trop 51(5):610–615. https://doi.org/10.1590/0037-8682-0174-2017
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN (2014) Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009 − 11 in 14 European and Mediterranean countries. J Antimicrob Chemother 69:1804–1814
CLSI (ed) (2009) Performance standards for antimicrobial disk susceptibility tests; Approved Standard-10th ed. CLSI documnet M02-A10. Clinical and laboratory Standards Institute, Pennsylvania
CLSI (ed) (2012) Methods for dilution antimicrobial suceptibility tests for bacteria that grow aerobically; Approved Standard-9th ed. CLSI document M07-A9 Clinical and laboratory Standards Institute, Pennsylvania
CLSI (2018) Performance standards for antimicrobial susceptibility testing; 28th ed Informational Supplement. CLSI documnet M100-S28. Clinical and laboratory Standards Institute, Pennsylvania
De Rosa A, Mutters NT, Mastroianni CM, Kaiser SJ, Günther F (2019) Distribution of carbapenem resistance mechanisms in clinical isolates of XDR Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 38:1547–1552. https://doi.org/10.1007/s10096-019-03585-0
Diab M, Fam N, El-Said M, El-Dabaa E, El-Defrawy I, Saber M (2013) Occurrence of VIM-2 Metallo-β-Lactamases in imipenem resistant and susceptible Pseudomonas aeruginosa clinical isolates from Egypt. Afr J Microbiol Res 7(35):4465–4472
Dogonchi AA, Ghaemi EA, Ardebili A, Yazdan-setad S, Pournajaf A (2018) Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: A potential threat to clinical therapeutics. Tzu Chi Med J 30(2):90–96
Engelkirk P, Duben-Engelkirk J (2008) Laboratory diagnosis of infectious diseases. Lippincott Williams and Wilkins, Philadelphia
Esmaeili D et al (2019) Alerting prevalence of MBLs producing Pseudomonas aeruginosa isolates. Gene Rep 16:100460
Feng W, Sun F, Wang Q et al (2017) Epidemiology and resistance characteristics of Pseudomonas aeruginosa isolates from the respiratory department of a hospital in China. J Glob Antimicrob Resist 8:142–147
Gaballah A, Elbaradei A, Elsheredy A, Kader O (2018) Emergence of blaVEB and blaGES among VIM producing Pseudomonas aeruginosa clinical isolates in Alexaneria, Egypt. Acta Microbiol Immunol Hung 66(1):131–142
Ismail SJ, Mahmoud SS (2018) First detection of New Dehli metallo-Beta-Lactamases variants (NDM-1, NDM-2) among Pseudomonas aeruginosa isolated from Iraqi hospitals. Iran J Microbiol 10(2):98–103
Jamal WY, Albert MJ, Rotimi VO (2016) High prevalence of New Delhi metallo-beta-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS One. https://doi.org/10.1371/e0152638
Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L (2011) NDM2 carbapenemase in Acinetobacter baumannii from Egypt. J Antimicrob Chemother 66:1260–1262
Kiddee A, Henghiranyawong K, Yimsabai J, Tiloklurs M, Niumsup PR (2013) Nosocomial spread of class 1 integron-carrying extensively drug-resistant Pseudomonas aeruginosa isolates in a Thai hospital. Int J Antimicrob Agents 42:301–306
Levy Hara G, Gould I, Endimiani A, Pardo PR, Daikos G, Hsueh PR, Mehtar S, Petrikkos G, Casellas JM, Daciuk L, Paciel D, Novelli A, Saginur R, Pryluka D, Medina J, Savio E (2013) Detection, treatment, and prevention of carbapenemase-producing Enterobacteriaceae: recommendations from an International Working Group. J Chemother 25(3):129–1S40
Li J, Zou M, Dou Q, Hu Y, Wang H, Yan Q et al (2016) Characterization of clinical extensively drug-resistant Pseudomonas aeruginosa in the Hunan province of China. Ann Clin Microbiol Antimicrob 15(1):35
Magiorakos AP, Srinivasan A, Carey RB et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281
Mahon CR, Lehman DC, Manuselis G (2014) Text Book Of Diagnostic Microbiology, 5th edn. W B Saunders, Missouri
Malkoçoğlu G, Aktaş E, Bayraktar B, Otlu B, Bulut ME (2017) VIM-1, VIM- 2, and GES-5 Carbapenemases among Pseudomonas aeruginosa isolates at a tertiary hospital in Istanbul, Turkey. Microb Drug Resist 23(3):328–334
Manzoor U, Siddique S, Ahmed R, Noreen Z, Bokhari H, Ahmad I (2016) Antibacterial, structural and optical characterization of mechano-chemically prepared ZnO nanoparticles. PLoS ONE. https://doi.org/10.1371/e0154704
Maroui I, Barguigua A, Aboulkacem A, Ouarrak K, Sbiti M, Louzi H, Timminouni M, Belhaj A (2016) First report of VIM-2 metallo-β-lactamases producing Pseudomonas aeruginosa isolates in Morocco. J Infect Chemother 22(3):127–132
Moore E, Arnscheidt A, Kruger A, Strompl C, Mau M (2004) Simplified protocols for the preparation of genomic DNA from bacterial cultures. In: Kowalchuk GA, Bruijn FJ, Head IM, Akkermans AD, Elsas JD (eds) Molecular Microbial Ecology Manual. Springer Netherlands, Dordrecht, pp 3–18
Oliver A, Mena A, Macià MD (2008) Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections. In: Baquero F, Nombela C, Cassell GH, Gutiérrez JA (eds) Evolutionary biology of bacterial and fungal pathogens. ASM Press ISBN 978-1-55581-414-4, pp 433–444
Patel G, Bonomo RA (2013) Stormy waters ahead: global emergence of carbapenemases. The multiple roles of antibiotics and antibiotic resistance in nature. Front Microbiol l4:48
Paul D, Dhar D, Maurya AP, Mishra S, Sharma GD, Chakravarty A et al (2016) Occurrence of co-existing blaVIM-2 and blaNDM-1 in clinical isolates of Pseudomonas aeruginosa from India. Ann Clin Microbiol Antimicrob 15:31
Pitout JD, Gregson DB, Poirel L, Mc Clure JA, Phillip L, Church DL (2005) Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J Clin Microbiol 43:3129–3135.
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123
Rizek C, Fu L, Dos Santos LC, Leite G, Ramos J, Rossi F, Guimaraes T, Levin AS, Costa SF (2014) Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann Clin Microbiol Antimicrob 13:43
Shaaban M, Al-Qahtani A, Al-Ahdal M, Barwa R (2017) Molecular characterization of resistance mechanisms in Pseudomonas aeruginosa isolates resistant to carbapenems. J Infect Dev Ctries 11:935–943
Shahat HS, Mohamed HMA, Abd Al-Azeem MW Nasef SA. (2019) Molecular Detection of Some Virulence Genes in Pseudomonas aeruginosa Isolated from Chicken Embryos and Broilers with regard to Disinfectant Resistance. SVU Int J Veter Sci 2(2):52–70
Shu JC, Chia JH, Siu LK, Kuo AJ, Huang SH et al (2012) Interplay between mutational and horizontally acquired resistance mechanisms and its association with carbapenem resistance amongst extensively drug-resistant Pseudomonas aeruginosa (XDR-PA). Int J Antimicrob Agents 39(3):217–222
Spilker T, Coenye T, Vandamme P, Lipuma JJ (2004) PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 42:2074–2079
Walsh F (2013) The multiple roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:255. https://doi.org/10.3389/fmicb.2013.00255
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR (2009) Characterization of a new metallo-β-lactamase gene, blaNDM-1,and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054
Zafer MM, Al-Agamy MH, El-Mahallawy HA, Amin MA, Ashour MS (2014b) Antimicrobial resistance pattern and their beta-lactamase encoding genes among Pseudomonas aeruginosa strains isolated from cancer patients. Biomed Res Int. https://doi.org/10.1155/2014/101635
Zafer MM, Amin M, El Mahallawy H, Ashour MS, Al Agamy M (2014a) First report of NDM-1-producing Pseudomonas aeruginosa in Egypt. Int J Infect Dis 29:80–81
Acknowledgements
Not applicable.
Funding
The expenses were paid by the authors.
Author information
Authors and Affiliations
Contributions
All authors proposed the research idea. AMB designed the plan of the work, performed the laboratory analysis, and shared in writing and submitting the manuscript. MIM wrote the protocol, managed the lab work, shared in writing the manuscript. GME validated the research experimental design, analyzed the data of this study, revised, and corrected the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Basha, A., El-Sherbiny, G.M. & Mabrouk, M.I. Phenotypic characterization of the Egyptian isolates “extensively drug-resistant Pseudomonas aeruginosa” and detection of their metallo-β-lactamases encoding genes. Bull Natl Res Cent 44, 117 (2020). https://doi.org/10.1186/s42269-020-00350-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-020-00350-8