Abstract
Background
Y-chromosome-specific short tandem repeat markers reside on the non-recombinant portion of the Y chromosome and are paternally inherited. These properties make Y-STRs a useful tool in investigations of sexual assault and other violent crimes, paternity cases, genealogical testing, and evolutionary studies. Native populations reside in the Peruvian coast, mountains, and jungle; however, the Mestizos are the largest population group in Peru.
Results
Of the 290 samples, 283 distinct haplotypes were observed. Gene diversities for each Y-STR marker ranged from 0.4271 to 0.8597. The haplotype diversity for the total sample was 0.9964.
Conclusions
Genetic affinity was identified between the Peruvian Mestizo population and other Peruvian populations (except Jivaro, Awajun, and Shipibo-Conibo populations) and Bolivian Mestizo populations using multidimensional scaling and phylogenetic tree analysis
Similar content being viewed by others
Background
Y-STR markers are inherited via the paternal lineage, present haploid, and except for the pseudoautosomal regions do not experience recombination. These properties make Y-STRs a useful tool in sexual assault investigations, paternity testing, genealogical testing, and evolutionary studies (Roewer 2009).
The populations of Peru have undergone different migratory processes, contributing to Peruvian population variation and native populations in the coast, mountains, and jungle as well as populations with substantial genetic admixture. Because of this diversity, studies have been carried out on autosomal STRs markers from different Peruvian populations (Delgado & Neyra 2018; Neyra et al. 2019, 2020; Neyra-Rivera et al. 2020; Neyra-Rivera et al. 2021a, b; Bermejo et al. 2022; Neyra-Rivera et al., 2022) and also on Y-STRs from native populations (Tineo et al. 2015; Neyra-Rivera et al., 2022; Neyra-Rivera et al. 2023) analysis in the case of Y-STRs the genetic diversity (GD, probability that two randomly chosen individuals can be differentiated by a particular genetic marker(s) (Nei 1987)), haplotype diversity (HD, probability that two randomly chosen individuals can be differentiated by a particular set of markers comprising a haplotype(s) (Nei 1987)), probability of a random match (MP, probability that two randomly selected individuals in a population share identical genotypes (Jones 1972)), discrimination capacity (DC, probability that two randomly selected individuals will be genetically differentiated at a particular marker(s) (Smalldon and Moffat 1973)), haploid match probability (HMP, probability that two randomly selected individuals will share the same haplotype), and pairwise genetic distance (RST, the correlation of allele sizes (rather than allelic states) between genetic markers sampled within populations (Excoffier 2001)). However, there have not been studies with large population samples (i.e., greater than 200 individuals) and with at most 23 Y-STR markers. Only one study with 83 individuals and 23 Y-STR markers (accession number YA005599) is represented in the YHRD database (Yhrd.org). The present investigation seeks to expand on available genetic data by studying a larger sample set of Mestizos for 27 Y-STR markers and compare these genetic data to assess relationships with populations associated either geographically or historically.
Methods
Samples and experiment
DNA from 290 unrelated individuals were collected in Nucleid Cards (Copan). The individuals reside in different departments of Peru (Amazonas n = 2, Ancash n = 15, Apurimac n = 4, Arequipa n = 15, Ayacucho n = 15, Cajamarca n = 2, Cusco n = 16, Huancavelica n = 4, Huanuco n = 22, Ica n = 4, Junín n = 9, La Libertad n = 2, Lambayeque n = 1, Lima n = 123, Loreto n = 4, Madre de Dios n = 7, Moquegua n = 1, Piura n = 5, Puno n = 31, Tacna n = 2, Tumbes n = 2, and Ucayali n = 4) (Fig. 1). All participants freely and voluntarily signed informed consent to participate in this study.
Departments of Peru from which the 290 samples were collected. (a) Amazonas n = 2, (b) Ancash n = 15, (c) Apurimac n = 4, (d) Arequipa n = 15, (e) Ayacucho n = 15, (f) Cajamarca n = 2, (g) Cusco n = 16, (h) Huancavelica n = 4, (i) Huanuco n = 22, (j) Ica n = 4, (k) Junín n = 9, (l) La Libertad n = 2, (m) Lambayeque n = 1, (n) Lima n = 123, (o) Loreto n = 4, (p) Madre de Dios n = 7, (q) Moquegua n = 1, (r) Piura n = 5, (s) Puno n = 31, (t) Tacna n = 2, (u) Tumbes n = 2, and (v) Ucayali n = 4
A 1.2 mm punch was taken from each card and amplified by direct PCR using the Yfiler ™ Plus PCR Amplification Kit (Life Technologies) following the manufacturer’s instructions. Amplified products were detected in the Applied Biosystem ™ 3500XL Genetic Analyzer (Life Technologies) following the manufacturer’s recommended protocol. After capillary electrophoresis, the data were imported into the GeneMapper® ID-X v1.5 software (Thermo Fisher Scientific 2015) to generate the genetic profiles which were analyzed according to the manufacturer’s recommendations. The population data were deposited in the YHRD database (accession number: YA006020).
Statistical analysis
For the calculation of the allelic frequencies for 23 of the 27 STRs, the Power Stats V1.2 software was used (Tereba 1919); for the DYS385a/b and DYF387S1 markers, the “type” frequencies were calculated by the counting method. GD and HD were calculated with the following formulas GD = (n/n − 1) (1 − ΣPἰ2) and HD = (n/n − 1) (1 − ΣXἰ2), where Pἰ is the frequency of the ἰth allele, n is the number of samples analyzed, and Xἰ represents the calculated frequency of the ἰth haplotype (Sandoval et al., 2013). The MP was calculated as MP = ΣXἰ2 and DC was determined by dividing the number of different observed haplotypes with the sum total number of identified haplotypes. HMP was calculated with the formula HMP = 1 − HD. AMOVA was used to RST values between Y-STR datasets. The RST values were then used for MDS analysis. RST and p values between different populations were estimated by AMOVA and visualized on a MDS plot (YHRD) (Willuweit & Roewer 2007).
Results
Allelic frequencies and forensic statistical parameter analyses
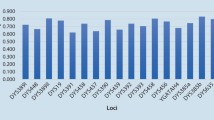
This study analyzed 27 Y STRs in 290 males collected across different departments in Peru. Results were obtained for all 27 Y-STR loci in all samples, and no null alleles were observed. Allele (or type) frequencies for each locus of the combined data set are shown in Table 1. The DYS437 marker displayed the fewest number of alleles (3 alleles), while the DYS385a/b marker had the highest number of types (45 types). GD values ranged between 0.4271 (DYS437) and 0.9415 (DYS385a/b), and only two Y-STR loci had values less than 0.5 (DYS437 and DYS391). The two highest diversity markers, based on GD, were DYS385a/b (0.9415) and DYF387S1 (0.9363) while the lowest diversity was observed at the DYS437 locus (0.4271).
Haplotype diversity
Since there is no recombination between the Y STRs, haplotype data better display the diversity within the population. There were 283 distinct haplotypes in the total data set (n = 290) with HD and DC values of 0.9964 and 0.9759, respectively (Table 2). The largest number of the same haplotype was two and seven pairs of two haplotypes were observed (H78 –Puno and Cajamarca-, H105 –Huancavelica and Cuso-, H124 –Lima-, H201 –Lima-, H210 –Lima-, H253 –Madre de Dios and Lima- and H255 –Lima and Cusco-). Ten microvariants were identified and were observed in the markers DYS627 (alleles 18.2, 20.2, and 21.2), DYS448 (alleles 16.2, 17.2, and 18.2), and DYS448 (allele 18.2).
Population structure for forensic analyzes
Using an online tool NevGen Y DNA Predictor (Cetkovic Gentula & Nevski, 2015) provisional haplogroups (most probable and major clade only) for the distinct haplotypes of the Mestizos from Peru were determined. In the Mestizo population, the majority (56.18%) belonged to haplogroup Q, with the rest distributed among haplogroups R (22.61%), J (7.07%), E (6.36%), G (2.83%), T (2.47%), I (2.12%), and L (0.35%) (Table 2).
Genetic distances
The RST values for genetic distances and the MDS plot are shown in Table 3 and Fig. 2 for the Mestizo population and other datasets from Boliviano (Bolivia), Mestizo (Bogotá, Colombia), Aymara (Puno, Peru), Ashaninka (Oxapampa, Peru), Awajun (Amazonas, Peru), Chachapoya (Amazonas, Peru), Chumbivilca (Cuzco, Peru), Chuquibamba (Arequipa, Peru), Huanca (Junín, Peru), Iquito (Loreto, Peru), Jivaro (Amazonas, Peru), Quechua (Lima, Peru), Shipibo-Conibo (Ucayali, Peru), and Spanish (Spain). There were 17 Y-STRs in common among the data sets (DYSDYS456, DYS389I, DYS390, DYS389II, DYS458, DYS19, DYS385, DYS393, DYS391, DYS439, DYS635, DYS392, YGATAH4, DYS437, DYS438, DYS448).
Multidimensional scaling (MDS) analysis for Mestizo population (n = 290) and 14 other sample populations. Bolivia [Boliviano] (n = 218)a; Bogotá, Colombia [Mestizo] (n = 184)b; Oxapampa, Peru [Ashaninka] (n = 57)c; Amazonas, Peru [Awajun] (n = 18)d; Puno, Peru [Aymara] (n = 90)e; Amazonas, Peru [Chachapoya] (n = 106)f; Cuzco, Peru [Chumbivilca] (n = 10)g; Arequipa, Peru [Chuquibamba] (n = 16)h; Junín, Peru [Huanca] (n = 13)i; Loreto, Peru [Iquito] (n = 85)j; Amazonas, Peru [Jivaro] (n = 21)k; Lima, Peru [Quechua] (n = 11)l; Ucayali, Peru [Shipibo-Conibo] (n = 21)m; and Spain [Spanish] (n = 2406).n deposited in the YHRD. Filed under accession numbers: a: YA005867, b: YP000047, c: YA005583, d: YA005891, e: YP000861, f: YA004108, g: YA003820, h: YA003821, i: YA003822, j: YP000860, k: YA005058, l: YA003949, m: YA003825, n: YP001017
Discussion
Molecular biology technologies offer enhanced capabilities for human identification. Forensic DNA typing has an important role in some criminal investigations, which in turn benefits society, by providing critical data for developing investigative leads to identify donors of biological evidence who may be perpetrators of crime as well as to exonerate those that may be wrongly associated with biological evidence. In order to contribute to assessing the significance of an association by DNA between a reference sample and an evidence sample, a population study on a sample population of Peruvians was carried out by analyzing 27 Y-STRs contained within a widely used commercial kit (i.e., Yfiler™ Plus PCR Amplification Kit).
The per locus GD of the studied population ranged between 0.4271 (DYS437) and 0.9415 (DYS385a/b). In contrast, the GD values for the Peruvian Aymara population were between 0.2424 (DYS437) and 0.9298 (DYS385a/b), Peruvian Ashaninca populations were between 0.1337 (DYS437)) and 0.6582 (DYS438) and the population of the Peruvian Jungle were between 0.000 (DYS635, DYS448 and DYS437) and 0.7094 (DYS449) (Tineo et al. 2015; Neyra-Rivera et al., 2022; Neyra-Rivera et al. 2023). The values have a similar distribution with those of the Mestizo population of Spain, DYS393 (0.4625) had the lowest GD (Martin et al., 2004).
Because there is no recombination on the Y chromosome where the Y-STRs reside, haplotype diversity is more meaningful than GD to compare diversity among populations. The Y-STR HD was 0.9964 with 283 distinct haplotypes out of 290 individuals analyzed in the Peruvian population studied herein. This level of diversity is slightly lower than that of the Peruvian Aymara (0.9998, n = 90) and Peruvian Asháninca (1.000) populations and higher than that of the Peruvian Jungle population (0.9910, n = 141) (Tineo et al. 2015; Neyra-Rivera et al., 2022; Neyra-Rivera et al. 2023). In addition, the diversity value was lower than that of the Mestizo populations of Spain (0.9995, n = 144), Colombia (0.9971, n = 137), and Bolivia (1.000, n = 100) (Martin et al., 2004; Vullo et al. 2015; Yunis et al. 2005). The difference compared with the Mestizo populations may be due in part to fewer samples being typed, a fewer number of markers tested (17 vs 27 markers), and/or stochastic variation.
The specific haplogroups carried by the males analyzed in the Peruvian Mestizo population provide insight into the male lineages of the population. The most prevalent was haplogroup Q (56.18%), which also was the most common haplogroup in the Ashaninca (between 99 and 100%), Aymara (86.52%), and La Selva (91.56%) populations. The lower proportion of haplogroup Q in the Mestizo population may be due to admixture that has occurred and continues to occur in Peru; for various reasons, native populations have migrated from their communities to the cities in Peru (Tineo et al. 2015; Neyra-Rivera et al., 2022; Neyra-Rivera et al. 2023).
The RST values were used to generate a UPGMA tree using the Molecular Evolutionary Genetics Analysis (MEGA) 6.06 software (Tamura et al. 2013). The Mestizo population was closest to Bolivian and other Peruvian populations (except Chachapoya and Quechua populations) (Fig. 3). The Mestizo population was closest to the Bolivian population (Bolivia, Boliviano) and farthest from the Jivaro and Awajun (Amazonas, Peru) and Shipibo-Conibo (Ucayali) (Fig. 2). These results suggest that there is a high genetic affinity between the Mestizo population and the other Peruvian populations (except Jivaro, Awajun and Shipibo-Conibo populations). Additionally, the Jivaro, Awajun, and Shipibo-Conibo populations were distant from all groups analyzed which may be due to genetic differences and/or small population sample sizes (n ≤ 21) (Fig. 3).
Conclusions
In conclusion, 27 Y-STR loci in 290 unrelated Mestizo males were typed, providing a larger dataset for this population group. The data obtained have been deposited in the YHRD database (accession number: YA006020). These data are larger than any other available data for the Mestizo population from Peru. For example, YRHD (Yhrd.org) contains 394 haplotypes (394 with 12 Y-STR haplotypes, 229 with 17 Y-STR haplotypes, 83 with 23 Y-STR haplotypes, and until the study herein 0 with 27 Y-STR haplotypes) (filed under accession numbers YA003399 and YA005599). A specific limitation of this study is the small sample sizes for some groups which can lead to errors in the allele frequency estimates and subsequent population genetic calculations. Regardless of this limitation, the analyses carried out herein show that the Y-STR haplotypes are highly polymorphic and thus can be useful in human identification forensic cases, with proper statistical methods that account for uncertainty, as well as may have value for studies on the history of the human populations in Latin America.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author.
Abbreviations
- AMOVA:
-
Analysis of molecular variance
- MDS:
-
Multidimensional scaling plot
- PCR:
-
Polymerase chain reaction
- YHRD:
-
Y Haplotype Reference Database
- Y-STR:
-
Y-chromosome-specific short tandem repeat
References
Bermejo ME, Ge J, Budowle B, Infante C, Neyra-Rivera CD (2022) Genetic study with 21 autosomal STRs in five Peruvian macro regions for human identification purposes. Legal Med 57:1344–6223. https://doi.org/10.1016/j.legalmed.2022.102073
Cetkovic M, Nevski A (2015). NevGen Y-DNA haplogroup predictor. In: http://www.nevgen.org/. Accessed 20 Feb 2023
Delgado E, Neyra CD (2018) Allele frequencies of 21 autosomal STR markers in a mixed race Peruvian population applied to forensic practice. Rev Espanola De Medicina Leg 45(3):92–97. https://doi.org/10.1016/j.reml.2018.09.002
Excoffier L (2001) Analysis of population subdivision. In: Balding DJ, Bishop M, Cannings C (eds) Statistical Genetics. John Wiley & Sons, Chichester, pp 271–324
Jones DA (1972) Blood lood samples- probalility of discrimination. J Forensic Sci Soc 12(2):355–359. https://doi.org/10.1016/s0015-7368(72)70695-7
Martín P, García-Hirschfeld J, García O, Gusmão L, García P, Albarrán C, Sancho M, Alonso A (2004) A Spanish population study of 17 Y-chromosome STR loci. Forensic Sci Int 139(2–3):231–235. https://doi.org/10.1016/j.forsciint.2003.11.012
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 39–46
Neyra CD, Delgado E, Robles C, Velasquez M, Caceres O, Budowle B (2020) Population genetic study of a Peruvian population using human identification STRs. Int J Legal Med 134(6):2071–2073. https://doi.org/10.1007/s00414-020-02418-6
Neyra CD, Suarez M, Cueva E, Bailón H, Gutierrez E (2019) Identificación genética de recién nacidos en Perú: un estudio piloto. Rev Chil Pediatr 90(1): 26–35. https://doi.org/10.32641/rchped.v90i1.730
Neyra-Rivera CD, Budowle B (2022) Genetic variation of different Peruvian populations using 23 autosomal STR markers. Forensic Sci Int Genet Suppl Ser 8:259–262. https://doi.org/10.1016/j.fsigss.2022.10.055
Neyra-Rivera CD, Delgado E, Diaz F, Quispe JS, Ge J, Budowle B (2021a) Genetic study with autosomal STR markers in people of the Peruvian jungle for human identification purposes. Can Soc Forensic Sci 54(3):117–138. https://doi.org/10.1080/00085030.2021.1933811
Neyra-Rivera CD, Robles CS, Delgado E, Velasquez MRE, Budowle B (2023) Analysis of 27 STR markers of the chromosome Y in the population of the jungle from Peru. Rev. Espanola de Medicina Leg ISSN 0377–4732. https://doi.org/10.1016/j.reml.2022.10.001
Neyra-Rivera CD, Ticona A, Delgado E, Velasquez MRE, Budowle B (2020) Allelic frequencies with 23 autosomic STRS in the Aymara population of Peru. Int J Legal Med 135(3):779–781. https://doi.org/10.1007/s00414-020-02448-0
Neyra-Rivera CD, Ticona A, Delgado E, Velasquez MRE, Caceres O, Budowle B (2021) Population data of 27 Y-chromosome STRS in Aymara population from Peru. Aust J Forensic Sci 1–15. https://doi.org/10.1080/00450618.2021.1882571
Roewer L (2009) Y chromosome STR typing in crime casework. Forensic Sci Med Pathol 5(2):77–84. https://doi.org/10.1007/s12024-009-9089-5
Sandoval JR, Lacerda DR, Jota MS, Salazar-Granara A, Vieira PP, Acosta O, Cuellar C, Revollo S, Fujita R, Santos FR, Genographic Project Consortium (2013) The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: the legacy of the Uros. PLoS ONE 8(9):e73006. https://doi.org/10.1371/journal.pone.0073006
Smalldon KW, Moffat AC (1973) The calculation of discriminating power for a series of correlated attributes. J Forensic Sci 13(4):291–295. https://doi.org/10.1016/S0015-7368(73)70828-8
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30 (12): 2725–2729. https://doi.org/10.1093/molbev/mst197
Tereba A (1919) Tools for analysis of population statistics. Profiles DNA 2:14–16
Thermo Fisher Scientific (2015). GeneMapper® iii ID-X Software v1.5 Reference Guide. In: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/100031707_GeneMapIDX_ver1_5_ReferenceGuide.pdf. Accessed 7 Febreury 2023
Tineo DH, Loiola S, Paredes FV, Noli LR, Amaya YC, Simãoc F, Carvalho EF, Gusmão L (2015) Genetic characterization of 27 Y-STR loci in the native population of Ashaninka from Peru. Forensic Sci Int Genet Suppl Ser 5:e220–e222. https://doi.org/10.1016/j.fsigss.2015.09.088
Vullo C, Gomes V, Romanini C, Oliveira AM, Rocabado O, Aquino J, Amorim A, Gusmão L (2015) Association between Y haplogroups and autosomal AIMs reveals intra-population substructure in Bolivian populations. Int J Legal Med 129(4):673–680. https://doi.org/10.1007/s00414-014-1025-x
Willuweit S, Roewer L (2007) Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet 1:83–87. https://doi.org/10.1016/j.fsigen.2007.01.017
Yunis JJ, Acevedo LE, Campo DS, Yunis EJ (2005) Population data of Y-STR minimal haplotypes in a sample of Caucasian-Mestizo and African descent individuals of Colombia. Forensic Sci Int 151(2–3):307–313. https://doi.org/10.1016/j.forsciint.2005.02.005
Acknowledgements
The authors thank the volunteers for donating their samples for the research. Also, to the Universidad Privada Peruano Alemana for financing this research.
Funding
We thank the Universidad Privada Peruano Alemana for financing the development of the research (RECTORAL RESOLUTION No. 013–2022-REC).
Author information
Authors and Affiliations
Contributions
Carlos Neyra, Edgardo Delgado, and Ericson Gutierrez participated in the design of the study. Carlos Neyra and Edgardo Delgado processed the samples and obtained the genetic profiles. Carlos Neyra, Edgardo Delgado, Ericson Gutierrez, and Bruce Budowle carried out the statistical analysis. All participated in the writing and approval of the final version of the article.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committee of Instituto de Medicina Tropical “Daniel Alcides Carrion” from San Marcos National University (Certificate of Approval CIEI-2018–015).
Consent for publication
Consent has been given by all authors for publication of this study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Neyra-Rivera, C.D., Ramos, E.D., Ingunza, E.G. et al. Analysis of 27 Y-chromosomal STR loci of the Mestizo Peruvian population. Egypt J Forensic Sci 13, 47 (2023). https://doi.org/10.1186/s41935-023-00366-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41935-023-00366-x