Abstract
Background
The COVID-19 pandemic has a large impact worldwide and is known to particularly affect the older population. This paper outlines the protocol for external validation of prognostic models predicting mortality risk after presentation with COVID-19 in the older population. These prognostic models were originally developed in an adult population and will be validated in an older population (≥ 70 years of age) in three healthcare settings: the hospital setting, the primary care setting, and the nursing home setting.
Methods
Based on a living systematic review of COVID-19 prediction models, we identified eight prognostic models predicting the risk of mortality in adults with a COVID-19 infection (five COVID-19 specific models: GAL-COVID-19 mortality, 4C Mortality Score, NEWS2 + model, Xie model, and Wang clinical model and three pre-existing prognostic scores: APACHE-II, CURB65, SOFA). These eight models will be validated in six different cohorts of the Dutch older population (three hospital cohorts, two primary care cohorts, and a nursing home cohort). All prognostic models will be validated in a hospital setting while the GAL-COVID-19 mortality model will be validated in hospital, primary care, and nursing home settings. The study will include individuals ≥ 70 years of age with a highly suspected or PCR-confirmed COVID-19 infection from March 2020 to December 2020 (and up to December 2021 in a sensitivity analysis). The predictive performance will be evaluated in terms of discrimination, calibration, and decision curves for each of the prognostic models in each cohort individually. For prognostic models with indications of miscalibration, an intercept update will be performed after which predictive performance will be re-evaluated.
Discussion
Insight into the performance of existing prognostic models in one of the most vulnerable populations clarifies the extent to which tailoring of COVID-19 prognostic models is needed when models are applied to the older population. Such insight will be important for possible future waves of the COVID-19 pandemic or future pandemics.
Similar content being viewed by others
Background
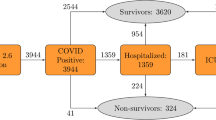
COVID-19 has a large impact worldwide, causing over 6 million deaths [1], various long-term health effects in a large group of individuals, and an increased burden on healthcare providers and medical institutions. Health outcomes of a COVID-19 infection can be severe, particularly in the older population [2]. In the Netherlands, it is estimated that disproportionately high mortality of 88.8% of all COVID-19 deaths occurred in the older population (≥ 70 years of age) even though they make up only 14% of the total population [3]. Similarly, these mortality proportions in older individuals were also relatively high in hospitalized (60%) and nursing home settings (40%) [3].
Hundreds of prognostic models have been developed to quantify (differences in) mortality risk or other outcomes in COVID-19 patients and to identify individuals at greater risk of developing various future health outcomes [4]. COVID-19 prognostic models have been used to facilitate informed shielding decisions by governments [5], identify higher-risk groups requiring ventilatory or critical care support early to enable targeted recruitment for randomized controlled trials [6], and deliver more personalized, risk-based treatments for which effectiveness is known to vary according to disease severity more precisely [7]. However, despite the development of hundreds of prognostic models, only a few models are of high quality and low risks of bias, according to a critical appraisal in a large living systematic review [4]. For those models that were appraised at a low risk of bias, information about their actual performance in external validation studies is scarce. Further, due to the added complexity of health conditions like frailty [8] and multimorbidity [9] in older individuals, we hypothesize that these prognostic models derived for the general adult population will underperform when validated in an older population.
In this protocol, we describe a comprehensive external validation study to evaluate the predictive performance of eight prognostic models in the older population, defined as individuals aged 70 and older, in hospital, primary care, and nursing home settings. The predictive performance of the prognostic models will be evaluated in a different population than they were derived on, being the older population of individuals aged 70 and older compared to the general adult population. One model will be evaluated in all healthcare settings (hospital care, primary care, and nursing home) to assess predictive performance across settings.
Methods
We have adhered to the TRIPOD guidelines checklist for external validation studies [10] in reporting this study protocol (Supplementary file 4).
Selection of COVID-19 prognostic models
In the living systematic review of diagnostic and prognostic prediction models for COVID-19 (www.covid-precise.org) [4], all published prediction models were reviewed using PROBAST (www.probast.org) [11, 12], a quality or risk of a bias assessment tool for prediction model studies. Using results from the fifth update of this review, we have identified all candidate prognostic models that predict the risk of mortality in individuals with COVID-19 infection with uncertain or low risk of bias. Fifteen candidate models met this criterion of which five prediction models (PRIEST [13], CUCAF-SF [14], CUCA-SF [14], and QCOVID [15] for males and females) were not included for validation due to the unavailability of data on certain predictors in the six cohorts of older patients while two prognostic scores (qSOFA [16] and NEWS [17]) were excluded because they express risk of mortality qualitatively rather than as a risk prediction (Supplementary file 1). Eight prognostic models were included for external validation (Fig. 1). Of these eight models, five were COVID-19-specific (GAL COVID-19 mortality model [18], 4C Mortality Score [19], NEWS2 + model [20], Xie model [21], and Wang clinical model [22]) and were developed in adult COVID-19 populations during the pandemic. Three prognostic models were already existing before COVID-19 pandemic and were used for the prediction of in-hospital mortality risk after admission for any respiratory infections or sepsis (APACHE-II [23], CURB65 [24], and SOFA [25]) (Table 1). The details of eight selected models can be found in Supplementary file 2.
Validation cohorts
Data for this external validation study is collected from six cohorts with older individuals presenting with COVID-19 infection in the Netherlands from three settings: a hospital setting (3 cohorts), primary care setting (2 cohorts), and nursing home setting (1 cohort) (Table 2).
Participants
The study participants consist of older individuals (≥ 70 years of age) presenting with highly suspected or reverse transcription polymerase chain reaction (RT-PCR) confirmed COVID-19 from March 2020 to December 2020 in hospital, primary care, and nursing home settings.
Before the widespread use of the RT-PCR test for COVID-19 diagnosis, participants were included using proxy criteria. In the hospital cohorts (CliniCo, COVID-OLD, COVID-PREDICT), the reported respiratory diseases that had COVID-like symptomology are used as an inclusion criterion until 31 March 2020. From April 2020 onwards, a confirmed RT-PCR test for COVID-19 is used as an inclusion criterion. In the primary care cohorts (PHARMO and JHN/ANH/AHA), the participants are included based on free text information and reports of respiratory infections. From June 2020 onwards, ICPC R83.03 for COVID-19 infection is used as an inclusion criterion. The nursing home cohort (Ysis) used RT-PCR as an inclusion criterion. Only participant data on the first presentation or admission of COVID-19 will be included. In the three hospital cohorts, admissions with a duration fewer than 7 days between discharge and readmission will be considered a single hospital admission.
Outcome
All prediction models have mortality as the predicted outcome (Table 1). In all three hospital cohorts, the outcome is defined as in-hospital mortality. In the primary care and nursing home cohorts, the outcome is defined as 28-day mortality.
Predictors
Definitions and timing of the predictor variables for the eight models were extracted from original publications (Table 1). Recorded variables in the cohorts are matched, as closely as possible, to the original predictor measurement procedures (Supplementary file 3).
Statistical analysis
We will externally validate the eight COVID-19 prognostic models in the six cohorts of older patients with COVID-19, aiming to assess their predictive performance when transported from a general adult population to a specific older population. The performance of the GAL-COVID-19 model is assessed across the three healthcare settings. The GAL-COVID-19 mortality model was developed in a primary care setting (general practitioners) and will be validated across different settings (in hospitals, primary care, and nursing homes) [26]. The 4C Mortality Score, NEWS2 + model, Xie model, Wang clinical model, APACHE-II score, CURB-65 score, and SOFA score were developed in hospitalized populations and will be externally validated in the same setting. Evaluation and assessment of the predictive performance of the COVID-19 prognostic models will be performed in each cohort separately. The statistical analysis will be performed in R (version 4.0.0 or later) [27].
Descriptive analysis
Participant characteristics and predictor information will be described in all study cohorts (overall and stratified on mortality outcome status) to identify differences in case-mix between the development and validation study populations [28]. These comparisons give insight into the expected model performance and transportability [26].
Missing data
The missing data will be described to determine possible reasons for and patterns in missingness [29]. Based on these findings, decisions about the handling of the missing values in the statistical analysis will be made. We anticipate that missing data will be handled using multiple imputations by chained equations using the Full Conditional Specification or Joint Modelling (JOMO) [30]. All variables and outcomes in the final prognostic models are included in the imputation model to ensure compatibility. A total of 50 imputed datasets will then be generated as cohorts are expected to have less than 50% missing values for all relevant variables [31].
Assessment of predictive performance
For each prognostic model, we will apply the model according to the authors’ original descriptions and evaluate its predictive performance. We will evaluate discrimination (the model’s ability to distinguish individuals who died after presentation with COVID-19 diagnosis from those who did not) and calibration (the agreement between predicted and observed mortality risks) in each cohort [32]. Discrimination will be assessed in all models by quantifying the area under the receiver operating characteristic curve, i.e., c-statistic [33], and pooled by taking the median c-statistic over imputed datasets and computing dispersion using the interquartile range [34].
Calibration will be assessed by visualizing the calibration of expected versus observed risk using LOESS-smoothed plots on stacked imputed data sets [35]. The GAL-COVID-19 mortality model, NEWS2 + model, Xie model, and Wang clinical model are model equations. For these models, calibration will be assessed in terms of the calibration-in-the-large coefficient and calibration slope [35]. The coefficients are again pooled on a log scale using Rubin’s rules in case of multiple imputed datasets [36]. For each performance measure, for each evaluated model, we will compute the point estimate, standard error, and 95% confidence interval.
Decision curve analysis
Decision curve analyses will be performed to quantify the net benefit achieved by each model for predicting the originally intended endpoint across a range of risk thresholds ranging from zero to one [37].
Updating
Prediction models showing miscalibration will be adjusted using an intercept update, and predictive performance will be re-assessed for the recalibrated model.
Sensitivity analysis
Two sensitivity analyses will be performed to assess the variation in the predictive performance of the eight COVID-19 prognostic models when implemented in different time periods: January 2021 to December 2021 and March 2020 to December 2021. Additionally, predictive performance in estimating the 90-day mortality risk will be evaluated in cohorts that have data available on this outcome (CliniCo, COVID-PREDICT, PHARMO, JHN/ANH/AHA, Ysis).
Sample size
Using national statistics from July 2020 to December 2021, the COVID-related mortality fraction for the older population (≥ 70 years) living at home in the Netherlands (including self-reported COVID-19-positive patients) was around 3% [3]. Although we expect higher mortality in nursing homes, hospitals, and ICU admissions, we will assume that an outcome incidence of 3% is the lowest event fraction that we will encounter for the current prediction model validation study. We take this fraction as a starting point for the sample size calculations, which are based on sample size calculation recommendations for external validation studies by Riley and colleagues [38]. Since we are considering various models, we base our sample size calculation around a general scenario that is reasonably applicable to all cases. Based on the distributions of predicted risk of mortality that were previously reported for some of the cohorts [8, 39], we assume the distribution of the linear predictor to be approximately N (− 3.9,1) [38]. Based on the calculations, a sample size of 754 will be required per cohort to validate an assumably well-calibrated model with a calibration-in-the-large coefficient of 0 and calibration slope of 1, and an assumed target standard error of calibration slope of 0.02. The target standard error of the calibration slope was chosen to ensure the evaluation of calibration with a higher precision than the typically used value of 0.05.
Discussion
This external validation study will assess the predictive performance of pre-existing clinical and COVID-19-specific prognostic models in the older population. Our study will reveal the validity of these COVID-19 prognostic models in one of the most vulnerable populations, the older population, as well as give insight into the requirements for tailored prediction models for the older population in future COVID-19 waves or pandemics.
While previous studies have largely focused on external validation of COVID-19 prediction models in the general adult population [14, 40, 41], this external validation study will focus on evaluating the performance of existing COVID-19 prognostic models specifically in the older population which represents the highest proportion of hospitalized COVID-19 patients and the highest mortality [3].
External validation in multiple cohorts across different healthcare settings allows for the assessment of between-setting heterogeneity in predictive performance and applicability at different points of care in older COVID-19 patients. This is one of the first studies to evaluate prognostic models for COVID-19 disease in individuals living in nursing homes.
Challenges and limitations
There are certain challenges that we anticipate encountering while conducting this research. Definitions of participant inclusion and predictor measurement procedures are expected to differ across cohorts. Although we have carefully planned to collect information on the definition of a COVID-19 infection and predictor measurements for each cohort, we cannot rule out the possibility of heterogeneity occurring across different settings and its effects on model performance [42]. However, this heterogeneity resembles practical conditions encountered in clinical practice and thus provides relevant knowledge on the anticipated performance of the models in clinical practice.
A limitation of this study is that not all low risk of bias COVID-19 prognostic models identified by the living systematic review could be included in this external validation. This was due to the lack of predictor information available in the cohorts. Similarly, predictor measurements and incidence of mortality due to COVID-19 can vary over time due to newer variants, better medical management, and improved vaccination coverage [43]. These temporal changes can potentially limit the predictive performance of the prognostic models being investigated in the current study.
Conclusion
External validation of newer and existing COVID-19 prognostic models can provide evidence about their predictive performance when implemented in the older population as tools of effective risk stratification and in aiding decision-making for targeted and timely clinical interventions.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to confidentiality. COVID-OLD, CliniCo, COVID-PREDICT, PHARMO, and JHN/ANH/AHA are available on reasonable request.
Abbreviations
- PCR:
-
Polymerase chain reaction
- APACHE score:
-
Acute Physiology and Chronic Health Evaluation score
- NEWS score:
-
National Early Warning Score
- SOFA score:
-
Sequential organ failure assessment score
- PROBAST:
-
Prediction model Risk of Bias Assessment Tool
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- JOMO:
-
Joint modeling
- LOESS:
-
Locally estimated scatterplot smoothing
References
WHO. WHO coronavirus (COVID-19) dashboard 2022. Overview. Available from: https://covid19.who.int.
Smorenberg A, Peters EJ, van Daele P, Nossent EJ, Muller M. How does SARS-CoV-2 targets the elderly patients? A review on potential mechanisms increasing disease severity. Eur J Intern Med. 2021;83:1–5.
Rijksoverheid. Sterfte 2022. Date accessed: April 2022. Available from: https://coronadashboard.rijksoverheid.nl/landelijk/sterfte.
Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369:m1328.
Smith GD, Spiegelhalter D. Shielding from COVID-19 should be stratified by risk. BMJ. 2020;369:m2063.
Furlow B. COVACTA trial raises questions about tocilizumab’s benefit in COVID-19. Lancet Rheumatol. 2020;2(10):e592.
Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, et al. Remdesivir for the treatment of COVID-19 - final report. N Engl J Med. 2020;383(19):1813–26.
Blomaard LC, van der Linden CMJ, van der Bol JM, Jansen SWM, Polinder-Bos HA, Willems HC, et al. Frailty is associated with in-hospital mortality in older hospitalised COVID-19 patients in the Netherlands: the COVID-OLD study. Age Ageing. 2021;50(3):631–40.
Barnett K, Mercer SW, Norbury M, Watt G, Wyke S, Guthrie B. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. Lancet. 2012;380(9836):37–43.
Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162(1):55–63.
Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1–33.
Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019;170(1):51–8.
Goodacre S, Thomas B, Sutton L, Burnsall M, Lee E, Bradburn M, et al. Derivation and validation of a clinical severity score for acutely ill adults with suspected COVID-19: the PRIEST observational cohort study. PLoS ONE. 2021;16(1):e0245840.
Bradley P, Frost F, Tharmaratnam K, Wootton DG, Research NWCOfR. Utility of established prognostic scores in COVID-19 hospital admissions: multicentre prospective evaluation of CURB-65, NEWS2 and qSOFA. BMJ Open Respir Res. 2020;7(1):e000729.
Clift AK, Coupland CAC, Keogh RH, Diaz-Ordaz K, Williamson E, Harrison EM, et al. Living risk prediction algorithm (QCOVID) for risk of hospital admission and mortality from coronavirus 19 in adults: national derivation and validation cohort study. BMJ. 2020;371:m3731.
Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, et al. Assessment of Clinical Criteria for Sepsis: for the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016;315(8):762–74.
Royal College of Physicians. National Early Warning Score (NEWS): standardising the assessment of acute- illness severity in the NHS. Report of a working party. London: RCP; 2012.
Gude-Sampedro F, Fernandez-Merino C, Ferreiro L, Lado-Baleato O, Espasandin-Dominguez J, Hervada X, et al. Development and validation of a prognostic model based on comorbidities to predict COVID-19 severity: a population-based study. Int J Epidemiol. 2021;50(1):64–74.
Knight SR, Ho A, Pius R, Buchan I, Carson G, Drake TM, et al. Risk stratification of patients admitted to hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339.
Carr E, Bendayan R, Bean D, Stammers M, Wang W, Zhang H, et al. Evaluation and improvement of the National Early Warning Score (NEWS2) for COVID-19: a multi-hospital study. BMC Med. 2021;19(1):23.
Xie J, Hungerford D, Chen H, Abrams ST, Li S, Wang G, et al. Development and external validation of a prognostic multivariable model on admission for hospitalized patients with COVID-19. 2020.
Wang K, Zuo P, Liu Y, Zhang M, Zhao X, Xie S, et al. Clinical and laboratory predictors of in-hospital mortality in patients with coronavirus disease-2019: a cohort study in Wuhan, China. Clin Infect Dis. 2020;71(16):2079–88.
Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818–29.
Lim WS, van der Eerden MM, Laing R, Boersma WG, Karalus N, Town GI, Lewis SA, Macfarlane JT. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58(5):377–82.
Vincent JL, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, Reinhart CK, Suter PM, Thijs LG. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22(7):707–10.
Justice AC, Covinsky KE, Berlin JA. Assessing the generalizability of prognostic information. Ann Intern Med. 1999;130(6):515–24.
Team Rc. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2022. Available from: https://www.r-project.org.
Debray TP, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol. 2015;68(3):279–89.
Donders AR, van der Heijden GJ, Stijnen T, Moons KG. Review: a gentle introduction to imputation of missing values. J Clin Epidemiol. 2006;59(10):1087–91.
James Carpenter MK. Multiple imputation and its application. 1st ed. 2012.
White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med. 2011;30(4):377–99.
Moons KG, Altman DG, Vergouwe Y, Royston P. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ. 2009;338:b606.
Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128–38.
Marshall A, Altman DG, Holder RL, Royston P. Combining estimates of interest in prognostic modelling studies after multiple imputation: current practice and guidelines. BMC Med Res Methodol. 2009;9:57.
Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW, Topic Group ’Evaluating diagnostic t, et al. Calibration: the Achilles heel of predictive analytics. BMC Med. 2019;17(1):230.
Rubin DB. Multiple imputation for nonresponse in surveys. New York: Wiley; 1987. https://doi.org/10.1002/9780470316696.
Vickers AJ, van Calster B, Steyerberg EW. A simple, step-by-step guide to interpreting decision curve analysis. Diagn Progn Res. 2019;3:18.
Riley RD, Debray TPA, Collins GS, Archer L, Ensor J, van Smeden M, et al. Minimum sample size for external validation of a clinical prediction model with a binary outcome. Stat Med. 2021;40(19):4230–51.
Rutten JJS, van Loon AM, van Kooten J, van Buul LW, Joling KJ, Smalbrugge M, et al. Clinical suspicion of COVID-19 in nursing home residents: symptoms and mortality risk factors. J Am Med Dir Assoc. 2020;21(12):1791–7.
Gupta RK, Marks M, Samuels THA, Luintel A, Rampling T, Chowdhury H, et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur Respir J. 2020;56(6):2003498.
de Jong VMT, Rousset RZ, Antonio-Villa NE, Buenen AG, Van Calster B, Bello-Chavolla OY, et al. Clinical prediction models for mortality in patients with covid-19: external validation and individual participant data meta-analysis. BMJ. 2022;378:e069881.
Luijken K, Groenwold RHH, Van Calster B, Steyerberg EW, van Smeden M. Impact of predictor measurement heterogeneity across settings on the performance of prediction models: a measurement error perspective. Stat Med. 2019;38(18):3444–59.
Watson OJ, Barnsley G, Toor J, Hogan AB, Winskill P, Ghani AC. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infectious Dis. 2022;22(9):1293–302. https://doi.org/10.1016/S1473-3099(22)00320-6.
Acknowledgements
We would like to acknowledge the COVID-19 outcomes in older people (COOP) consortium investigators: Evertine J. Abbink, Wilco P. Achterberg, Sifra H. van de Beek, Marian Beekman, Ludo F.M Beenen, Jesse M. van den Berg, Marieke T. Blom, Bram van den Borst, Sebastiaan J.H. Bredie, Frederiek van den Bos, Virgil A.S.H Dalm, Yvonne M. Drewes, Carline J. van den Dries, Petra J.M. Elders, Miriam C. Faes, Jan Festen, Geert-Jan Geersing, Jacobijn Gussekloo, Miriam L. Haaksma, Vanessa C. Harris, Ron M.C Herings, Cees M.P.M Hertogh, Jacobien J Hoogerwerf, Jeannette Jacobs-Peters, Steffy Jansen, Karlijn J. Joling, Anneke G.Julien, Veerle M.G.T.H van der Klei, Anna Kuranova, P. Hugo M. van der Kuy, Carolien M.J van der Linden, Anouk M. van Loon, Kim Luijken, Josephine S. van de Maat, Francesco U.S Mattace Raso, René J.F. Melis, Julia Minnema, Simon P. Mooijaart, Dennis O. Mook-Kanamori, Karel G.M Moons, Mihai G. Netea, Geeske Peeters, Harmke A. Polinder-Bos, Bas F.M van Raaij, Roos S.G Sablerolles, P. Eline Slagboom, Maarten van Smeden, Rosalinde A.L. Smits, Annemieke Smorenberg, Lisanne Tap, Hannah M. la Roi-Teeuw, Lisa S. van Tol, Hanna C. Willems, and Anum Zahra. We are grateful to the older people advisory board for their helpful feedback and insight as representatives of the older persons during this project.
Funding
This study is supported by grants from the Netherlands Organization for Scientific Research (ZonMW, grant number: 10430102110005).
Author information
Authors and Affiliations
Consortia
Contributions
AZ performed the methodology, investigation, writing—original draft, and visualization. KL performed the conceptualization, methodology, investigation, writing—review and editing, and supervision. EA and JP performed the writing—review and editing, and data curation for a hospital cohort. JvdB and MTB performed the investigation, writing—review and editing, and data curation for the primary care data. PE, JF, and JG performed the writing—review and editing. KJ performed the writing—review and editing, and data curation for a nursing home cohort. RM performed the investigation and writing—review and editing, and performed the data curation for a hospital cohort. SM performed the project administration and funding acquisition. HPB performed the investigation and writing—review and editing, and performed the data curation for a hospital cohort. BvR performed the investigation and writing—review and editing, and data curation for a hospital cohort. AS performed the investigation and writing—review and editing, and data curation for a hospital cohort. HT performed the investigation and writing—review and editing, and data curation for the primary care data. CM performed the conceptualization, methodology, and writing—review and editing, and supervision. MvS performed the conceptualization, methodology, and writing—review and editing, supervision, and project administration. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study will be conducted in accordance with the EU GDPR (General Data Protection Regulation). Due to the fact that this study does not require direct patient or physician involvement, the need for formal ethical reviewing was waived by the local medical research ethics committee (MREC Utrecht, the Netherlands), according to Dutch law.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary file 1.
Description of excluded prognostic models.
Additional file 2: Supplementary file 2.
Prediction models and risk scores.
Additional file 3: Supplementary file 3.
Predictor measurements in derivation and validation cohorts.
Additional file 4: Supplementary file 4.
TRIPOD checklist for validation of prediction model
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zahra, A., Luijken, K., Abbink, E.J. et al. A study protocol of external validation of eight COVID-19 prognostic models for predicting mortality risk in older populations in a hospital, primary care, and nursing home setting. Diagn Progn Res 7, 8 (2023). https://doi.org/10.1186/s41512-023-00144-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41512-023-00144-2