Abstract
Introduction
Pneumonia is a leading cause of death in Malaysia. Whilst many studies have reported the aetiology of pneumonia in Western countries, the epidemiology of pneumonia in Malaysia remains poorly understood. As carriage is a prerequisite for disease, we sought to improve our understanding of the carriage and antimicrobial resistance (AMR) of respiratory tract pathogens in Malaysia. The rural communities of Sarawak are an understudied part of the Malaysian population and were the focus of this study, allowing us to gain a better understanding of bacterial epidemiology in this population.
Methods
A population-based survey of bacterial carriage was undertaken in participants of all ages from rural communities in Sarawak, Malaysia. Nasopharyngeal, nasal, mouth and oropharyngeal swabs were taken. Bacteria were isolated from each swab and identified by culture-based methods and antimicrobial susceptibility testing conducted by disk diffusion or E test.
Results
140 participants were recruited from five rural communities. Klebsiella pneumoniae was most commonly isolated from participants (30.0%), followed by Staphylococcus aureus (20.7%), Streptococcus pneumoniae (10.7%), Haemophilus influenzae (9.3%), Moraxella catarrhalis (6.4%), Pseudomonas aeruginosa (6.4%) and Neisseria meningitidis (5.0%). Of the 21 S. pneumoniae isolated, 33.3 and 14.3% were serotypes included in the 13 valent PCV (PCV13) and 10 valent PCV (PCV10) respectively. 33.8% of all species were resistant to at least one antibiotic, however all bacterial species except S. pneumoniae were susceptible to at least one type of antibiotic.
Conclusion
To our knowledge, this is the first bacterial carriage study undertaken in East Malaysia. We provide valuable and timely data regarding the epidemiology and AMR of respiratory pathogens commonly associated with pneumonia. Further surveillance in Malaysia is necessary to monitor changes in the carriage prevalence of upper respiratory tract pathogens and the emergence of AMR, particularly as PCV is added to the National Immunisation Programme (NIP).
Similar content being viewed by others
Highlights
-
Bacterial carriage in the upper respiratory tract in Malaysia is poorly understood.
-
Klebsiella pneumoniae was the most prevalent bacteria isolated.
-
33.8% of isolates were resistant to at least one antibiotic.
Introduction
Lower respiratory tract infections, including pneumonia, are the sixth leading cause of death across all ages globally, and the primary cause of mortality in children aged 4 years and under [1]. In Malaysia, pneumonia was the second highest cause of death across all ages in 2017 [2]. Community acquired pneumonia (CAP) is associated with a 4.2% fatality rate [3], which is lower than rates published in Western countries (7.7–12%) [4, 5]. This is possibly due to less-stringent reporting in Malaysia and therefore underestimates of mortality.
Whilst many studies define the aetiology of pneumonia in Western countries, its epidemiology in Malaysia is less understood. Globally S. pneumoniae is reported as the most common cause of bacterial pneumonia [6]. Pneumococcal conjugate vaccines (PCVs) offer protection against serotypes of S. pneumoniae most commonly associated with disease or antimicrobial resistance (AMR), and are included in the national immunisation programmes (NIPs) of 144 countries worldwide [7]. As of June 2020, Malaysia, Thailand, Sri Lanka, China and Vietnam were yet to include PCV in their NIPs [8]; however the Malaysian government has announced its intention to implement PCV as part of their NIP from late 2020 [9]. PCV vaccine types (VT) 19F, 14, 6B, 1 and 19A are a major cause of invasive pneumococcal disease (IPD) in Malaysia, whilst 19F, 23F, 14, 6B, 1 and 3 are the most prevalent causes in South East Asia [10], therefore PCV implementation is of likely benefit for the reduction of IPD and other diseases including pneumococcal pneumonia.
In addition to S. pneumoniae, multi-drug resistant ‘ESKAPE’ pathogens such as K. pneumoniae have been reported as common causative organisms of CAP [11, 12]. In contrast to Western countries, studies from Asia suggest K. pneumoniae is a more prominent cause of pneumonia than S. pneumoniae and H. influenzae, [13,14,15] with data from Malaysia suggesting K. pneumoniae to be the most prominent cause [16]. Like S. pneumoniae, the World Health Organization considers K. pneumoniae a priority AMR pathogen in need of control. Prevention strategies however are hindered by its high level of strain diversity [17]. K. pneumoniae is therefore a pathogen of concern. Whilst there is currently no vaccine for K. pneumoniae, disease due to H. influenzae type b (Hib) has been successfully reduced through the implementation of the Hib vaccine. Despite Hib vaccine implementation and high coverage in Malaysia [18], H. influenzae (including Hib) is still a major source of disease such as pneumonia and is a main cause of meningitis [16, 19].
As carriage is a prerequisite for disease [20], we sought to improve our understanding of carriage prevalence and AMR profiles of common upper respiratory tract (URT) pathogens in Malaysia, particularly those associated with pneumonia. This was done by undertaking a population-based carriage study. In particular, we hoped to investigate an understudied part of the Malaysian population (rural communities of Sarawak).
Methods
Study site
Sarawak, Sabah and Labuan comprise East Malaysia; also known as Malaysian Borneo due to its location on the island of Borneo. Sarawak is the largest state in Malaysia. The population consists of more than 40 ethnic subgroups; such as Malay, Malaysian Chinese and numerous native communities (commonly referred to as Dayak) comprising Orang Ulu (including Penan, Kelabit and Kenyah), Iban, Bidayuh, Melanau, Tagalm, Punan Bah, Kedayan and Suluk [21]. Many native communities live according to tribal traditions, residing in villages consisting of longhouses, with farming a common occupation. Such communities can be isolated due to their location, restricting access to modern medicines and healthcare services. However, it is becoming increasingly common for some residents to commute, with some staying in larger cities during the working week or longer, for work and study. This offers improved access to healthcare. Furthermore, governmental health initiatives such as a monthly flying doctor and Komuniti Sihat Pembina Negara (KOSPEN) improve public health in such communities by increasing access to medical professionals and raising awareness through the use of community health volunteers.
Five rural communities were sampled (Fig. 1); two were longhouse communities (Rumah Numpang and Rumah Bana) located in Sebauh, Bintulu, Sarawak. Although only 30-40 km from Bintulu town, travel to Rumah Numpang required a 20–30 min longboat ride up the Sungai Pandan and Sungai Sujan rivers to reach an isolated community within a clearing in forest land. Rumah Bana, an affluent community, was less isolated located 20-30 km from Bintulu town off the main road from Sebauh town. The remaining communities (Long Kerangan, Ba Marong and Long Nen) were very isolated, located in Ulu Baram, Miri, Sarawak. Long Kerangan was a highly deprived longhouse community, being isolated and impoverished, located 80 km from Long Lama (the nearest town and healthcare provider) in the middle of forest land. Ba Marong was an even more isolated longhouse community, located in the middle of dense forest 157 km from Long Lama. Travel to this site required trucks with four-wheel drive due to the tough terrain and lack of roads. Long Nen, a village rather than a longhouse community, was situated within dense forest 99 km from Long Lama. The rural communities sampled as part of this study encompassed different tribal lifestyles and included people of Iban, Kelabit, Kenyah and Penan ethnicity.
Map of recruitment sites.© Copyright 2020 | Multiplottr.com | All Rights Reserved. a Rumah Bana, Sebauh, Bintulu, Sarawak - GPS co-ordinate 3.142234, 113.279135; 43 participants recruited (all Iban). b Rumah Numpang, Sebauh, Bintulu, Sarawak - GPS co-ordinate 3.139950, 113.418036; 41 participant recruited (39 Iban, 1 unknown, 1 Penan). c Long Kerangan, Ulu Baram, Miri, Sarawak - GPS co-ordinate 3.766312, 114.841122; 19 participants recruited (all Penan). d Ba Marong, Ulu Baram, Miri, Sarawak - GPS co-ordinate 3.680444, 115.014777; 13 participant recruited (11 Penan, 1 Kelabit, 1 Kenyah). e Long Nen, Ulu Baram, Miri, Sarawak - GPS co-ordinate 3.679871, 114.897062; 24 participants recruited (all Penan)
Recruitment
A population-based survey of bacterial carriage was undertaken in participants of all ages from five rural communities in Sarawak, Malaysia during April 2016. Rural communities were identified by local community health workers and charities. Prior to each survey, local researchers sent a letter to/approached each longhouse elder/community lead to provide a study overview and seek approval for the team to visit. Researchers visited communities whose elder/lead gave approval and anyone present at each location were approached and invited to participate. Translators and community health workers facilitated recruitment and sampling.
Sample collection and transport
Bacterial carriage of the URT was sampled by taking whole mouth, nasal, oropharyngeal (OP) and nasopharyngeal (NP) swabs from each participant. Paediatric NP samples were taken using rayon tipped Transwab® Pernasal Amies with charcoal (Medical Wire and Equipment, Corsham, UK). Adult NP samples and all nasal and OP samples were taken using the appropriate viscose tipped sterile Amies swabs with charcoal (Deltalab, Chalgrove, UK). After collection, swabs were chilled where possible before being transported by World Courier at ambient temperature to the University of Southampton, UK. Transportation took 3–5 days.
Questionnaire
Participants were asked to complete a questionnaire requesting basic demographic data. Parents/guardians of child participants were responsible for the completion of questionnaires.
Assessment of bacterial carriage
Upon arrival at the laboratory, each swab was immersed and vortexed in skim milk, tryptone, glucose and glycerine (STGG) storage media. For each swab, 10 μL was plated onto Columbia blood agar with horse blood (CBA, Oxoid, UK), Columbia blood agar with chocolated horse blood (CHOC, Oxoid, UK), Columbia Blood Agar with Colisitin and Naladixic Acid (CNA, Oxoid, UK), Columbia Agar with Chocolated Horse Blood and Bacitracin (BACH, Oxoid, UK) and Lysed GC Selective Agar (GC, Oxoid, UK). Plates were incubated for 24–48 h at 37 °C in 5% CO2 and examined for the presence of Moraxella catarrhalis, S. pneumoniae, H. influenzae, Neisseria meningitidis, Staphylococcus aureus and K. pneumoniae. For the isolation of Pseudomonas aeruginosa, 10 μL was plated onto Pseudomonas CFC Selective agar (CFC, Oxoid, UK) and incubated for 24–48 h at 37 °C. Primary identification of all bacterial species was by colonial morphology. Bacteria of interest were then confirmed (Supplementary Methods 1) and isolates frozen at − 80 °C in STGG.
Pneumococcal serotyping
S. pneumoniae were serotyped by slide agglutination reactions using a Neufeld S. pneumoniae antisera kit (Statens Serum Institute, Copenhagen, Denmark).
Antibiotic susceptibility
Bacterial isolates were phenotypically tested for antibiotic resistance using MRSA agar and antibiotic discs and/or minimum inhibitory concentration (MIC) strips, in accordance to EUCAST (Supplementary Methods 1).
Carriage prevalence
Prevalence of carriage was calculated as a percentage by dividing the number of bacteria isolated by the total number of participants swabbed. This was done for each bacterial species according to anatomical sample site and age group. True carriage, defined as identification of a bacterial species in any specimen from an individual participant regardless of the site or number of carriage sites per person, was also determined.
Statistical analysis
The Fisher’s exact test was used to determine the significance of association between an independent and dependent variable (e.g. bacterial carriage and age group). A p-value of 0.05 was used as a significant threshold. All data analysis was done using GraphPad Prism version 7.03 for Windows (GraphPad Software, San Diego, CA, USA).
Results
In total 140 participants were recruited from five communities. Grouped by age, 8.6% (n = 12) of participants were 0–4 years, 20.0% (n = 28) were 5–17 years, 70.7% (n = 99) were 18 years and above and 0.7% (n = 1) were of unknown age. 44.3% (n = 62) of participants were male and 54.3% (n = 76) were female, whilst 1.4% (n = 2) of participants did not provide their gender. Regarding ethnicity, 58.6% (n = 82) of participants were Iban, 0.7% (n = 1) Kelabit, 0.7% (n = 1) Kenyah, 39.3% (n = 55) Penan participants and 0.7% (n = 1) did not provide ethnicity. All 140 participants provided mouth and nasal swab samples; 93.6% (n = 131) provided NP and 92.1% (n = 129) provided OP swab samples.
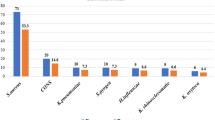
Prevalence of bacterial carriage was determined by anatomical sample site (Fig. 2). The organism most commonly isolated from the oropharynx and mouth was K. pneumoniae with a carriage prevalence of 27.1% (95CI: 19.7–35.74%; n = 35) and 12.9% (95CI: 7.8–19.6%; n = 18) respectively. For the nasopharynx and nose however, S. aureus was most prevalent at 13.7% (95CI: 8.4–20.8%; n = 18) and 12.1% (95CI: 7.2–18.7%; n = 17) respectively. All other species of interest showed much lower carriage levels at each of the anatomical sites sampled. True carriage was also determined (Fig. 3). K. pneumoniae was the most prevalent bacteria with a true carriage prevalence of 30.0% (95CI: 22.6–38.3%; n = 42), followed by S. aureus at 20.7% (95CI: 14.3–28.4%; n = 29), S. pneumoniae at 10.7% (95CI: 6.1–17.1%; n = 15), H. influenzae at 9.3% (95CI: 5.0–15.4%; n = 13), M. catarrhalis at 6.4% (95CI: 3.0–11.9%; n = 9), P. aeruginosa at 6.4% (95CI: 3.0–11.9%; n = 9) and N. meningitidis at 5.0% (95CI: 2.0–10.0%; n = 7).
True bacterial carriage of each bacteria by age group. Prevalence bar plot showing true bacterial carriage prevalence by age in participants recruited from rural communities in Sarawak, Malaysia in April 2016. True carriage is defined as at least one carriage event observed regardless of the site of isolation or the number of sites from which the bacterium was recovered. Error bars show standard error
When age group is considered, S. pneumoniae were carried significantly more in children aged 0–4 years (P < 0.05) (Fig. 3), and there were significantly more H. influenzae isolated from those aged 5–17 years than children under 5 years and adults (P < 0.05). K. pneumoniae were carried significantly more in adults than children (P < 0.05).
Of the 21 pneumococci isolated, 33.3% (n = 7) were PCV13 VT (6B, 14, 18C and 3), 14.3% (n = 3) were PCV10 VT (6B, 14 and 18C) and 28.6% (n = 6) were non-vaccine types (NVT) (6C, 7C, 10A, 11C and 35A) (Fig. 4). The final 38.1% (n = 8) could not be serotyped.
Serotypes of the S. pneumoniae isolated. Bar plot showing the observed numbers for each serotype identified for the 21 pneumococcal isolates collected from the oropharynx, nasopharynx, nose and mouth of participants recruited from rural communities in Sarawak, Malaysia in April 2016. ND: Not determined. PCV: pneumococcal conjugate vaccine, NVT: Non-vaccine type. Both serotype 3 S. pneumoniae were isolated from the same participant, one from the nasopharynx the other from the oropharynx. Two of the serotype 7C S. pneumoniae were isolated from the same participant, one from the nasopharynx the other from the nose. Both serotype 10A S. pneumoniae were isolated from the same participant, one from the nasopharynx the other from the nose. 6 of the un-typed S. pneumoniae were isolated from 3 participants with each having S. pneumoniae isolated from the nasopharynx and nose
All S. aureus isolates were identified as methicillin sensitive. When assessing antibiotic susceptibility of bacterial isolates using antibiotic discs and MIC strips, susceptibility ranged from 33.3–100% dependant on species and antibiotic (Fig. 5). With the exception of S. pneumoniae, all isolates of each bacterial species were susceptible to at least one type of antibiotic (e.g. all H. influenzae isolates were susceptible to ciprofloxacin), whilst all bacterial isolates, no matter the species, were susceptible to one or more of the antibiotics they were tested against. Of the 21 S. pneumoniae isolated, 47.6% (n = 10) were resistant to at least one antibiotic and 23.8% (n = 5) were resistant to two antibiotics. None were resistant to three or more antibiotics. Of the 19 H. influenzae isolated, 52.6% (n = 10) were resistant to at least one antibiotic and none were resistant to two or more antibiotics. Of the 42 S. aureus isolated, 66.7% (n = 28) were resistant to at least one antibiotic, 28.6% (n = 12) were resistant to at least two antibiotics and 2.3% (n = 1) were resistant to three antibiotics. Of the 17 M. catarrhalis isolated none were resistant to any antibiotic tested. Of the 58 K. pneumoniae isolated, 8.6% (n = 5) were resistant to at least one antibiotic and none were resistant to two or more antibiotics. Overall, 33.8% (n = 53) of all bacteria isolated and tested (n = 157) were resistant to at least one antibiotic, 10.8% (n = 17) to at least two antibiotics and 0.6% (n = 1) to three antibiotics. The highest prevalence of resistance was observed for S. aureus with 66.7% resistant to benzylpenicillin. This was followed by H. influenzae resistance to benzylpenicillin (52.6%) and S. pneumoniae to erythromycin (28.6%).
Antimicrobial resistance of bacteria isolated. Percentage bar plot showing the AMR of all 157 S. pneumoniae, H. influenzae, K. pneumoniae, S. aureus and M. catarrhalis isolates collected from residents of rural communities in Sarawak, Malaysia in April 2016. Antibiotic discs and MIC strips were used and EUCAST breakpoints were used to assess susceptibility and resistance
Discussion
This study aimed to address the paucity of published epidemiological data in Malaysia, by increasing our knowledge of the carriage and AMR profiles of common URT pathogens. It has provided important data regarding bacterial epidemiology in an understudied part of the Malaysian population. As well as an invaluable perspective as to the potential challenges of infection and AMR control in such populations, which are isolated to varying degrees from the wider Malaysian population and/or are socio-economically distinct from those living in large towns or Peninsular Malaysia due to their rural or longhouse lifestyles. In Malaysia the epidemiology of URT pathobionts is poorly understood and would therefore benefit from further large-scale population-based studies to inform vaccine policy, to monitor the effects of vaccine implementation and provide an understanding of the prevalence of AMR to inform treatment of infection [22].
This study provided a particular opportunity to investigate ESKAPE pathogens such as K. pneumoniae, whose morbidity and mortality has been linked to an increased emergence of AMR [12]. Further surveillance is required to monitor changes in carriage prevalence, the emergence of new serotypes and AMR [23]. We found K. pneumoniae to be carried significantly more in adults, which is comparable to findings from Indonesia [24], however carriage of K. pneumoniae here was more than double that previously reported in Indonesia, Vietnam, Brazil and Minnesota, USA (between 3 and 15%) [24,25,26,27]. This is noteworthy because carriage is a risk factor for infection and K. pneumoniae is a common cause of CAP and serious sequela such as sepsis and meningitis [25, 28]. K. pneumoniae is of increasing concern due to the emergence of multidrug-resistant strains associated with hospital outbreaks and severe community-acquired infections. Last-line defense antibiotics are quickly becoming ineffective [29]. Despite this, isolates obtained in this study showed low prevalence of AMR, and none were multidrug resistant. Whilst only four antibiotics were tested, those chosen are the most clinically relevant based on prescription guidelines in Malaysia. Even though both interest and published clinical data for K. pneumoniae has increased, little has been published on the AMR of carriage isolates. A previous study found 29% of URT carriage isolates to have resistance to two of the antibiotics tested, with 100% of isolates being resistant to ampicillin [30]. Despite K. pneumoniae being considered an extended-spectrum beta-lactamase (ESBL)-producing bacteria [31], high levels of antibiotic susceptibility were found, even to β-lactam antibiotics [30], which is consistent with the findings of this study.
Carriage of S. aureus was consistent with the 23.4% carriage prevalence previously found in Peninsular Malaysia [32]. Due to a lack of data, the carriage prevalence of S. pneumoniae, H. influenzae and M. catarrhalis in Malaysia is unclear. Carriage data from young children in Indonesia has shown 49.5, 27.5 and 42.7% prevalence for S. pneumoniae, H. influenzae and M. catarrhalis respectively [33]. Due to the small sample size of this age group in our study, the carriage prevalence of these bacteria is imprecise; 50% (95CI: 21.8–78.2%), 0% (95CI: 0.0–26.5%), and 25% (95CI: 0.60–49.4%) for S. pneumoniae, H. influenzae and M. catarrhalis respectively. Further research would be useful in all age groups.
Whilst our study found 33.3% of carried S. pneumoniae serotypes to be PCV13 VT and 14.3% PCV10 VT, studies from Peninsula Malaysia showed a higher proportion of S. pneumoniae VT carriage, with up to 69.5% being PCV13 VT and 40.6% being PCV10 and PCV7 VT [34, 35]. Certainly, VT S. pneumoniae cause the majority of pneumococcal disease in Peninsular Malaysia with 66.3% of RTI in children being caused by PCV13 VT and 60% by PCV10 VT [36]. As PCV13 serotypes 1, 6B, 14, 19F, 19A are commonly associated with disease in Malaysia [10, 37], the carriage of 6B and 14 (which was in children) here is of interest for public health, particularly as these serotypes are included in both PCV10 and PCV13. However, since only 21 isolates of S. pneumoniae were recovered, limited conclusions can be drawn, especially as many remained un-typed. However, as PCV coverage of invasive isolates in Malaysia has been reported to be as high 88.2% for PCV13 and 64.1% for PCV10 the widespread uptake of PCV as part of the planned NIP update has a high potential to reduce disease [10, 38].
AMR is an increasing issue across Asia; multidrug resistant S. pneumoniae and K. pneumoniae are of particular concern [17, 36, 39, 40]. In Malaysia, antibiotics are commonly prescribed for the treatment of URT infections, although inappropriate antibiotic choice and dosage have been shown in up to 40% of cases. This is important, as the misuse of antibiotics is a significant selective force for the development of AMR. Erythromycin and penicillin are the most commonly prescribed and the most commonly over or incorrectly prescribed [41]. They were also the antibiotics shown to have the highest levels of resistance in this study. Furthermore, data suggests 55% of Malaysians discontinue their course of antibiotics when symptoms disappear, rather than completing the full course as prescribed. Such misuse is a significant risk factor for the development of AMR and is most common in community rather than hospital settings [42]. To reduce antibiotic misuse, education on the risks of AMR is being provided in accordance to the Malaysian Action Plan on Antimicrobial Resistance (MyAP-AMR) [23, 42].
Our AMR data contrasts with previous studies in Peninsular Malaysia; here 66.7% (n = 28) of S. aureus were resistant to benzylpenicillin compared to 100% seen previously [43]. Erythromycin resistance in S. pneumoniae was comparable to that from disease in Peninsular Malaysia study (28.6% versus 35.8%) whilst tetracycline resistance was lower (19.1% versus 38.7%) [44].
The main limitation of this study was that limited recruitment, especially in children, meant some carriage prevalence had wide confidence intervals. Low child recruitment was exacerbated by not all children providing NP or OP swabs. Of the 40 children recruited, only 32 provided NP swabs and 29 provided OP swabs; all provided nose and mouth swabs. Several factors accounted for low recruitment. In Rumah Numpang and Rumah Bana a high proportion of the community (particularly males) live away for work/study and only return on alternate weekends, whilst others live away for months at a time. Furthermore, some residents avoided areas that the study team were authorised to recruit from to avoid interaction with researchers/taking part in the study (Long Nen), whilst other sites were very small communities (Ba Marong comprised only two extended families). This study does however inform the feasibility of undertaking such research. Ways to improve recruitment such as the need to visit more sites or make repeat visits to sites have been identified. Although difficult for some communities, particularly those most isolated, authors would recommend increased community engagement prior to the research visit. Despite the University having links to the some of the communities and engaging with the leaders prior to the study, increased engagement may enable increased trust from community residents, reducing avoidance of researchers. It may also allow researcher to have better awareness of the best times to visit when most residents are likely to be present.
Limitations arose during the transportation of swabs, firstly due to temperature fluctuations between sample collection and arrival at UK laboratories because of unrefrigerated transportation conditions. During recruitment and sampling, utilised swabs were stored in insulated boxes with ice to keep temperatures low; however, temperatures reached up to 30 °C by arrival at excursion headquarters. At excursion headquarters and during ground courier transportation, swabs were kept in air-conditioned buildings and vans so temperatures remained below 21 °C. On the flight from Malaysia to the UK, the temperature was much lower (less than 10 °C), whilst ground transportation in the UK likely remained between 2 and 15 °C. Despite this, the temperature range of swab storage was compliant with the quality control conditions tested under Clinical Laboratory Standards Institute (CLSI) Approved Standard M40-A2 [45]. All swabs used in this study were CLSI Approved Standard M40-A2, which ensures bacterial numbers will not reduce by more than 3 log10 under at refrigeration and ambient temperature (2–30 °C) or increase by more than 1 log10 at refrigeration temperature, during a 48 h holding period. The second limitation regarding swab transportation, was the time between sampling and the swabs being processed in the laboratory. As transportation took 3–5 days, there is the potential that the carriage prevelances seen here may be underestimates due to some bacteria not surviving transportation. Certainly, transport conditions would have reduced bacterial viability, and may have affected some less resilient species more than others. Future studies would benefit from the use of dry shippers, which would combat the issue of temperature fluctuations and transportation delays. Furthermore, it may be of benefit to immediately store swab samples in STGG whilst in the field, rather than waiting until swabs are received by the laboratory. This would further facilitate freezing/use of dry shippers prior to transportation.
The need to transport swabs to UK based laboratories resulted from a lack of capacity at the time for microbial analyses at local collaborating institutions. This advocates building research and laboratory capacity in Malaysia.
Since whole genome sequencing was not possible in this study, many (38.1%, n = 8) S. pneumoniae remained un-typed; it is unclear whether they were non-typeable S. pneumoniae (lacking a capsule), were novel serotypes, or typical serotypes that were unable to be typed with the agglutination methodology used. Such data would have contributed to further understanding the potential benefit of widespread uptake in PCV in Malaysia and thus will be included in future research.
Conclusion
This study reports novel data on an understudied part of the Malaysian population, providing important insights into the epidemiology and AMR profiles of common respiratory pathogens associated with pneumonia.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Global Burden of Disease 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1151–210.
Statistics on Causes of Death, Malaysia, 2018 [press release]. Malaysia: Department of Statistics Malaysia, 31st October 2018 2018.
Azmi S, Aljunid SM, Maimaiti N, Ali A-A, Muhammad Nur A, De Rosas-Valera M, et al. Assessing the burden of pneumonia using administrative data from Malaysia, Indonesia, and the Philippines. Int J Infect Dis. 2016;49:87–93. https://doi.org/10.1016/j.ijid.2016.05.021.
Garcia-Vidal C, Fernandez-Sabe N, Carratala J, Diaz V, Verdaguer R, Dorca J, et al. Early mortality in patients with community-acquired pneumonia: causes and risk factors. Eur Respir J. 2008;32(3):733–9. https://doi.org/10.1183/09031936.00128107.
Limper AH. 97 - overview of pneumonia. In: Goldman L, Schafer AI, editors. Goldman's Cecil medicine (twenty Fourth Edition). Philadelphia: W.B. Saunders; 2012. p. 587–96. https://doi.org/10.1016/B978-1-4377-1604-7.00097-X.
GBD 2016 Lower Respiratory Infections Collaborators, et al. Lancet Infect Dis. 2018;18(11):1191–210.
World Health Organization. WHO-UNICEF estimates of PCV3 coverage 2019 [cited 2020 01/03/2020]. Available from: http://apps.who.int/immunization_monitoring/globalsummary/timeseries/tswucoveragepcv3.html.
World Health Organization and Immunization Vaccines and Biologicals Database. Vaccine in National Immunization Programme Update. 2020 [updated June 2020; cited 2020 18/06/2020]. Available from: http://www.who.int/immunization/monitoring_surveillance/VaccineIntroStatus.pptx.
Ko Chung Sen, DAP Malaysia. Budget 2020 achieved universal pneumococcal vaccination at 10% of the cost 2019 [Available from: https://dapmalaysia.org/statements/2019/10/13/29124/.
Jauneikaite E, Jefferies JM, Hibberd ML, Clarke SC. Prevalence of Streptococcus pneumoniae serotypes causing invasive and non-invasive disease in South East Asia: a review. Vaccine. 2012;30(24):3503–14. https://doi.org/10.1016/j.vaccine.2012.03.066.
Cilloniz C, Dominedo C, Torres A. Multidrug resistant gram-negative Bacteria in community-acquired pneumonia. Crit Care. 2019;23(1):79. https://doi.org/10.1186/s13054-019-2371-3.
McNeil HC, Lean SS, Lim V, Clarke SC. The state of ESKAPE in Malaysia. Int J Antimicrob Agents. 2016;48(5):578–9. https://doi.org/10.1016/j.ijantimicag.2016.08.011.
Chalmers J, Campling J, Ellsbury G, Hawkey PM, Madhava H, Slack M. Community-acquired pneumonia in the United Kingdom: a call to action. Pneumonia (Nathan Qld). 2017;9:15.
Prina E, Ranzani OT, Torres A. Community-acquired pneumonia. Lancet. 2015;386(9998):1097–108. https://doi.org/10.1016/S0140-6736(15)60733-4.
Chung DR, Song JH, Kim SH, Thamlikitkul V, Huang SG, Wang H, et al. High prevalence of multidrug-resistant nonfermenters in hospital-acquired pneumonia in Asia. Am J Respir Crit Care Med. 2011;184(12):1409–17. https://doi.org/10.1164/rccm.201102-0349OC.
Liam CK, Lim KH, Wong CM. Community-acquired pneumonia in patients requiring hospitalization. Respirology. 2001;6(3):259–64. https://doi.org/10.1046/j.1440-1843.2001.00336.x.
Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh CN, Dance DAB, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from south and Southeast Asia. Genome Med. 2020;12(1):11. https://doi.org/10.1186/s13073-019-0706-y.
World Health Organization. WHO-UNICEF estimates of Hib3 coverage 2019 [cited 2020 01/03/2020]. Available from: http://apps.who.int/immunization_monitoring/globalsummary/timeseries/tswucoveragehib3.html.
McNeil HC, Jefferies JM, Clarke SC. Vaccine preventable meningitis in Malaysia: epidemiology and management. Expert Rev Anti-Infect Ther. 2015;13(6):705–14. https://doi.org/10.1586/14787210.2015.1033401.
Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, O'Brien KL, et al. The fundamental link between pneumococcal carriage and disease. Expert Review of Vaccines. 2012;11(7):841–55. https://doi.org/10.1586/erv.12.53.
Vincent JR, Ali RM. Managing natural wealth : environment and development in Malaysia. Washington, D.C.: Singapore: RFF Press ; Institute of Southeast ASian Studies (ISEAS); 2005.
McNeil HC, Clarke SC. Serotype prevalence of Streptococcus pneumoniae in Malaysia - the need for carriage studies. Med J Malaysia. 2016;71(3):134–8.
Ministry of Health Malaysia. Malaysian Action Plan on Antimicrobial Resistance (MyAP-AMR) 2017-2021. Infection control unit, Medical Care Quality Section, Medical Development Division, Ministry of Health Malaysia, vol. 2018. p. 20–34.
Farida H, Severin JA, Gasem MH, Keuter M, van den Broek P, Hermans PW, et al. Nasopharyngeal carriage of Klebsiella pneumoniae and other gram-negative bacilli in pneumonia-prone age groups in Semarang, Indonesia. J Clin Microbiol. 2013;51(5):1614–6. https://doi.org/10.1128/JCM.00589-13.
Dao TT, Liebenthal D, Tran TK, Ngoc Thi Vu B, Ngoc Thi Nguyen D, Thi Tran HK, Thi Nguyen CK, Thi Vu HL, Fox A, Horby P, van Nguyen K, Wertheim HFL Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PLoS One 2014;9(3):e91999, https://doi.org/10.1371/journal.pone.0091999.
Lima ABM, Leão LSNdO, Oliveira LSdC, Pimenta FC. Nasopharyngeal gram-negative bacilli colonization in Brazilian children attending day-care centers. Braz J Microbiol. 2010;41(1):24–7. https://doi.org/10.1590/S1517-83822010000100005.
Wolf B, Rey LC, Moreira LB, Milatovic D, Fleer A, Verhoef J, et al. Carriage of gram-negative bacilli in young Brazilian children with community-acquired pneumonia. Int J Infect Dis. 2001;5(3):155–9. https://doi.org/10.1016/S1201-9712(01)90091-8.
Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–61. https://doi.org/10.1128/MMBR.00078-15.
Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–81. https://doi.org/10.1073/pnas.1501049112.
Gorrie CL, Mirceta M, Wick RR, Judd LM, Wyres KL, Thomson NR, et al. Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to Acquisition in the Referring Hospital. Clin Infect Dis. 2018;67(2):161–70. https://doi.org/10.1093/cid/ciy027.
Ferreira RL, BCM d S, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front Microbiol. 2019;9(3198).
Choi CS, Yin CS, Bakar AA, Sakewi Z, Naing NN, Jamal F, et al. Nasal carriage of Staphylococcus aureus among healthy adults. J Microbiol Immunol Infect. 2006;39(6):458–64.
Dunne EM, Murad C, Sudigdoadi S, Fadlyana E, Tarigan R, Indriyani SAK, et al. Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: a cross-sectional study. PLoS One. 2018;13(4):e0195098. https://doi.org/10.1371/journal.pone.0195098.
Yatim MM, Masri SN, Desa MNM, Taib NM, Nordin SA, Jamal F. Determination of phenotypes and pneumococcal surface protein a family types of Streptococcus pneumoniae from Malaysian healthy children. J Microbiol Immunol Infect. 2013;46(3):180–6. https://doi.org/10.1016/j.jmii.2012.04.004.
Cleary DW, Morris DE, Anderson RA, Alattraqchi AG, Rahman NIA, Ismail S, et al. The upper respiratory tract microbiome of indigenous Orang Asli in north-eastern Peninsular Malaysia. medRxiv. 2020;2020:06.02.20120444.
Goh SL, Kee BP, Abdul Jabar K, Chua KH, Nathan AM, Bruyne J, et al. Molecular detection and genotypic characterisation of Streptococcus pneumoniae isolated from children in Malaysia. Pathog Glob Health. 2020:1–9.
Subramaniam P, Jabar KA, Kee BP, Chong CW, Nathan AM, de Bruyne J, et al. Serotypes & penicillin susceptibility of Streptococcus pneumoniae isolated from children admitted to a tertiary teaching hospital in Malaysia. Indian J Med Res. 2018;148(2):225–31. https://doi.org/10.4103/ijmr.IJMR_1987_16.
Arushothy R, Ahmad N, Amran F, Hashim R, Samsudin N, Azih CRC. Pneumococcal serotype distribution and antibiotic susceptibility in Malaysia: a four-year study (2014-2017) on invasive paediatric isolates. Int J Infect Dis. 2019;80:129–33. https://doi.org/10.1016/j.ijid.2018.12.009.
Song J, Jung S, Ko K, K N, son J, Chang H, et al. high prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob Agents Chemother. 2004;48(6):2101–7. https://doi.org/10.1128/AAC.48.6.2101-2107.2004.
Mustafa M, Yusof I, Rahman M, Sein M, Tan T. Health care-associated pneumonia: pathogenesis diagnosis and preventions. IOSR J Pharm. 2014;4(12):47–52.
Tan G-H, Low Q-W, Lim H-C, Seah H-K, Chan H-K. Inappropriate antibiotic utilization: outpatient prescription review of a regional secondary Hospital in Kedah, Malaysia. J Pharm Pract Community Med. 2017;3(4):215–9. https://doi.org/10.5530/jppcm.2017.4.62.
Fatokun O. Exploring antibiotic use and practices in a Malaysian community. Int J Clin Pharm. 2014;36(3):564–9. https://doi.org/10.1007/s11096-014-9937-6.
Zarizal S, Yeo CC, Faizal GM, Chew CH, Zakaria ZA, Jamil Al-Obaidi MM, et al. Nasal colonisation, antimicrobial susceptibility and genotypic pattern of Staphylococcus aureus among agricultural biotechnology students in Besut, Terengganu, east coast of Malaysia. Tropical Med Int Health. 2018;23(8):905–13. https://doi.org/10.1111/tmi.13090.
Yasin RM, Zin NM, Hussin A, Nawi SH, Hanapiah SM, Wahab ZA, et al. Current trend of pneumococcal serotypes distribution and antibiotic susceptibility pattern in Malaysian hospitals. Vaccine. 2011;29(34):5688–93. https://doi.org/10.1016/j.vaccine.2011.06.004.
Clinical Laboratory Standards Institute. Quality control of microbiology transport systems; approved standard - second edition. CLSI document M40-A2. Wayne, PA: Clinical and Laboratory Standards Institute; 2014.
Acknowledgements
The authors thank Prof Mak Joon Wah for his scientific contribution and support.
We thank Winston Yap, Ann Yap, Jason Chin, Victor Lim, Susiana Chelz Macha and Audrey Yap for their assistance with transportation, recruitment, sample taking and translation.
We thank the Microbiology Department at Queen Alexandra Hospital Portsmouth for undertaking MALDI-TOF to confirm suspected K. pneumoniae isolates.
We also thank each community and their elder/community lead for allowing us to visit. We especially thank those who participated in the study.
Funding
This work was funded by a Newton Fund Institutional Links award to Dr. Stuart C Clarke and Prof Mohd Nor Norazmi [grant number 172686537]; the International Medical University [grant number BP I-01/12 (09) 2015].
Author information
Authors and Affiliations
Consortia
Contributions
DEM - formal analysis, investigation, data curation, writing of original draft, review & editing, visualisation and project administration. HM - investigation, data curation, review & editing, supervision and project administration. REH - formal analysis, data curation, writing of original draft and visualisation. RA - investigation, data curation, review & editing and project administration. ACT - investigation and review & editing. ST - investigation, data curation and review & editing. MNN - methodology, writing - review & editing and funding acquisition. VL - methodology, review & editing and funding acquisition. TCS - methodology, review & editing and funding acquisition. PKCL - review & editing and funding acquisition. CCW - methodology, investigation, review & editing and funding acquisition. DWC - investigation, review & editing and supervision. IKSY - methodology, investigation, resources, review & editing, supervision, project administration and funding acquisition. SCC - conceptualisation, methodology, resources, review & editing, supervision, project administration and funding acquisition. MYCarriage group - review & editing. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was provided by the International Medical University Malaysia (IMU) Joint-Committee on Research and Ethics (Project ID No: IMU R 175/2015) and the University of Southampton Faculty of Medicine Ethics Committee (Submission ID: 14478). Facilitated by translators throughout, a process for gaining informed consent was followed for all participants, with parents/guardians providing consent for those under 18 years. Participation involved reading and understanding the study information sheet and the completion of a consent form and questionnaire. In cases of poor literacy, study paperwork was read to participants, and consent forms signed to the best of the participant’s ability with support from literate family members.
Competing interests
SCC acts as principal investigator on studies conducted on behalf of University Hospital Southampton NHS Foundation Trust/University of Southampton that are sponsored by vaccine manufacturers but receives no personal payments from them. SCC has participated in advisory boards for vaccine manufacturers but receives no personal payments for this work. SCC has received financial assistance from vaccine manufacturers to attend conferences. DWC was a post-doctoral researcher on projects funded by Pfizer and GSK between April 2014 and October 2017. All grants and honoraria are paid into accounts within the respective NHS Trusts or Universities, or to independent charities. All other authors have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary Methods 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morris, D.E., McNeil, H., Hocknell, R.E. et al. Carriage of upper respiratory tract pathogens in rural communities of Sarawak, Malaysian Borneo. Pneumonia 13, 6 (2021). https://doi.org/10.1186/s41479-021-00084-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41479-021-00084-9