Abstract
Background
The Antarctic minke whale, Balaenoptera bonaerensis, is a Southern Hemisphere species of balaenopterid whale generally found south of 60°S in austral summer. In the Atlantic Ocean, they migrate north during austral winter as far as approximately 7°S. On 05 February, 2013, a 7.7 m baleen whale was observed floating dead off of Iberia Parish, Louisiana, USA in the northern Gulf of Mexico.
Results
Genetic analysis of mitochondrial and nuclear DNA sequences obtained from this animal determined that it was an Antarctic minke whale, Balaenoptera bonaerensis.
Conclusion
This is the first record of an Antarctic minke whale in the Gulf of Mexico.
Similar content being viewed by others
Background
The cetacean fauna of the northern Gulf of Mexico is comprised of tropical and subtropical species. Twenty-one cetacean species, twenty of which are odontocetes, are seen regularly (Mullin et al. 1991; Mullin & Fulling 2004) and are thought to have year-round resident populations in the Gulf of Mexico. Only 1 mysticete or baleen whale species, the Bryde’s whale Balaenoptera edeni, is known to have a resident population in the Gulf of Mexico, and this population appears to be phylogenetically distinct from all other Bryde’s whale populations worldwide (Rosel & Wilcox 2014). Other baleen whale species, including humpback, fin, sei and common minke, have been recorded in the Gulf of Mexico, but sightings, or strandings, are only occasional and these species are thought to be uncommon to rare in occurrence in this region (Jefferson & Schiro 1997).
One species that has not yet been documented in the northern Gulf of Mexico is the Antarctic minke whale, Balaenoptera bonaerensis Burmeister, 1867. Minke whales are the smallest of the balaenopterid whales and two species of minke whales are recognized, the common minke whale, B. acutorostrata, and the Antarctic minke whale. The common minke whale is found throughout temperate waters of the Northern and Southern Hemispheres from 65°S to 80°N (Perrin & Brownell 2009), including the North Atlantic where it is commonly found north of 40°N in both the eastern and western North Atlantic. It is rare in the Gulf of Mexico, however, and considered an accidental visitor to this region. The Antarctic minke whale is considered a Southern Hemisphere endemic (Deméré 2014) and is found throughout the Southern Hemisphere, generally south of 60°S in austral summer. This species undertakes seasonal migrations northward during the austral winter and in the western Atlantic the northernmost known wintering area is off the coast of Brazil to approximately 7°S (Perrin & Brownell 2009; Rice 1998). Here we report on the first record of a Southern Hemisphere minke whale species in the northern Gulf of Mexico. The whale stranded dead in Louisiana and genetic analysis of mitochondrial DNA and nuclear gene sequences were used to identify it to species.
Results and discussion
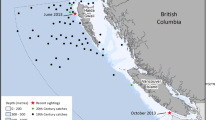
Mitochondrial and nuclear DNA sequences were successfully obtained from an unidentified dead baleen whale, 7.7 m in length that was found near Lake Point in Marsh Island Refuge, Iberia Parish, Louisiana, USA (29.55°, −91.72°; Figs. 1 & 2, Additional file 1: Table S1) in 2013.
Antarctic minke whale, Balaenoptera bonaerensis Burmeister, 1867, stranding location at Marsh Island, Iberia Parish, Louisiana, USA (red star) as well as recent Northern Hemisphere records for this species in the boreal North Atlantic, Togo, and Suriname. Yellow circles denote animals identified as hybrids by Glover et al. 2010; Glover et al. 2013
Phylogenetic analyses of 344 base pairs (bp) of the mitochondrial DNA (mtDNA) control region [GenBank Accession KU215790] placed the unknown stranded whale from the northern Gulf of Mexico firmly in the B. bonaerensis clade (Fig. 3) using both MrBayes and DNA Surveillance (posterior probability = 1.0, bootstrap value =100 %). Application of DNA Surveillance to the cytb sequence [GenBank Accession KU215791] returned the same result (data not shown).
Bayesian phylogenetic reconstruction of mtDNA control region sequences to identify species of UNKNOWN stranded balaenopterid whale, Balaenoptera bonaerensis Burmeister, 1867. Numbers at nodes present the posterior probabilities for nodes with values less than 0.9. All unlabeled nodes had posterior probabilities greater than 0.90. Tips are labeled with abbreviated species names and GenBank accession numbers. Species abbreviations: Bbon Balaenoptera bonaerensis; Bacu B. acutorostrata; Bomu B. omurai; Bede B. edeni; Bbor B. borealis; Bede_GOMx B. edeni from the Gulf of Mexico; Bphy B. physalus; B. mus B. musculus; Mnov Megaptera novaeangliae
A total of 1963 bp, 2278 bp, and 2277 bp of DNA sequence spanning four nuclear loci was obtained for the unidentified baleen whale, and for B. acutorostrata and B. edeni samples, respectively [GenBank Accessions KU215771-KU215789]. The total sequence length was shorter for the unidentified whale due to an indel in one allele of the CAT locus, rendering downstream sequence unreadable. After the addition of available GenBank sequences (Additional file 1: Table S2; B. bonaerensis sequences were not available in Genbank for INT), each alignment was truncated to the shortest sequence length resulting in final alignments of 430 bp, 294 bp, 508 bp, and 663 bp for AMBN exon 13, CAT, INT and LALBA, respectively. We identified a total of 58 variable sites and four 1 bp indels and one 21 bp indel within the truncated, aligned 1895 bp of nuclear data when comparing the three balaenopterid species (Fig. 4). For CAT, the unknown baleen whale sequence was identical to seven GenBank entries for known B. bonaerensis samples from Jackson et al. (Jackson et al 2009) and no fixed differences or indels were present in comparison to the other available Antarctic minke whale sequences (Fig. 4). On the other hand, there were three fixed differences and a single base pair indel between the unknown whale and the 13 B. acutorostrata samples at this locus. This animal exhibited from 0 (INT) to 12 (AMBN exon 13) fixed differences from common minke whales and a 21 bp deletion in INT further distinguishing it from common minke whales and Gulf of Mexico Bryde’s whales. Overall, in comparison to the common minke whale and the Gulf of Mexico Bryde’s whale, the unknown stranded whale exhibited a total of 16 and 19 fixed differences, respectively, across the four loci (Fig. 4).
In combination, the mitochondrial and nuclear DNA sequence data indicate that the whale that stranded in the northern Gulf of Mexico was indeed an Antarctic minke whale and further that it was not a hybrid of the two minke whale species. Multiple species-specific nucleotide differences and indels were found between the unknown stranded whale and B. acutorostrata and B. edeni nuclear sequences. The strongest evidence comes from CAT where the unknown whale exhibited sequence identical to seven Antarctic minke whales available in GenBank and from INT where the unknown whale had a 21 bp deletion when compared to the common minke whale (14 individuals). If the stranded animal were a hybrid with a common minke whale, it should have exhibited the longer and shorter alleles, which would have rendered the sequence unreadable at the point of the indel. This was not the case.
This stranding represents the first record for an Antarctic minke whale in U.S. waters of the western North Atlantic and GOMx. In general, common minke whales are uncommon in the GOMx. (Jefferson and Schiro 1997) present 10 records they deemed reliable after thoroughly examining stranding data and observer data. All records are ascribed to the common minke whale, all are strandings and the majority are from winter months. Jefferson and Schiro 1997) and Mitchell (1991) suggested that the winter pattern of strandings is indicative of strays during northward migration from lower-latitude breeding grounds. This explanation cannot account for the presence of this Antarctic minke whale in the Northern Hemisphere as during February, the austral summer, this species is found on its feeding grounds in the Antarctic southward of 60°S latitude. The stranded whale was a female and at 7.7 m in length is slightly smaller than expected for a sexually mature female of either the Antarctic minke whale or the common minke whale in the North Atlantic, and slightly longer than expected for a sexually mature female common minke whale from the Southern Hemisphere (Deméré 2014). Photos of the skull also support that the stranded whale was a minke whale (Fig. 5).
Although a very surprising result, this animal is not the first record for Antarctic minke whale in the Northern Hemisphere (Fig. 1). Rice 1998) reported a specimen from the Atlantic coast of South America in Suriname. More recently, Glover et al. (2010) identified a male Antarctic minke whale north of the Arctic Circle in the eastern North Atlantic using genetic analysis. Finally, a relatively small (~6 m) whale was entangled in purse seine net and brought to shore in the Gulf of Guinea, Togo where it was identified as a juvenile Antarctic minke whale based on morphological characteristics (Segniagbeto et al. 2014).
Recently, minke whale hybrids have been detected in the eastern North Atlantic based on genetic analysis (Glover et al. 2010; Glover et al. 2013). Interestingly, the direction of hybridization was different in the two cases. One female whale captured during the Norwegian commercial harvest in 2007 was the product of a female Antarctic minke whale and male B. acutorostrata, most likely of the North Atlantic subspecies B. acutorostrata acutorostrata, but which subspecies was not fully resolved (Glover et al. 2010). The second whale was also a female, taken in 2010. This whale was identified as the product of a female B. acutorostrata acutorostrata and male Antarctic minke whale; it was also pregnant, suggesting these hybrids are capable of reproducing, and the fetus was determined to be a backcross into B. acutorostrata acutorostrata (Glover et al. 2013).
Photos taken of the stranded whale from the Gulf of Mexico indicate a solid white ventral side with throat grooves that do not extend to the umbilicus. However, much of the epidermis of the animal was abraded away and therefore the complete color pattern is difficult to deduce. No photos of the baleen, a diagnostic characteristic used to separate the two minke whale species, were available. Photos of the pectoral fin, a feature that can be useful for distinguishing among minke whale species and subspecies, do not show evidence for the bright white flipper patch distinctive of the common minke whale in the North Atlantic (Deméré 2014), further supporting the diagnosis of an Antarctic minke whale. The pure Antarctic minke whale from the Northeast Atlantic identified by Glover et al. 2010) also lacked the flipper patch as expected, but at least one of the 2 hybrids did not differ morphologically from that expected from B. acutorostrata acutorostrata, i.e., it did exhibit the flipper patch (Glover et al. 2010). The ventral side of the flipper of the stranded whale appears to be solid white. The dorsal side is dark with a white trailing edge. White also appears on the body above the insertion of the pectoral fin, a characteristic reminiscent of the dwarf minke whale, an unnamed subspecies that is also restricted to the Southern Hemisphere.
One unanswered question across these sightings is why Antarctic minke whales are so recently being found in the Northern Hemisphere. Perhaps there has always been a low level of ‘leakage’ of Antarctic minke whales into the Northern Hemisphere. Increased interest and effort to recover and identify stranded and bycaught animals coupled with advances in molecular genetic techniques for species identification may simply now be better able to reveal the presence of the species in the Northern Hemisphere. Alternatively, could climate change and the coincident changes in the Antarctic ecosystem play a role, as postulated by Glover et al. (2013)? Whatever the cause, the increase in documented cases of inter-ocean movements has relevance for understanding and tracking future changes in distributions and habitat usage, as well as tracing of disease outbreaks in cetaceans. Cross-hemisphere movements of whales have generally been thought to be a rare event and transfer of a disease epidemic across the equator rarely considered. Continued examination of recovered whale carcasses coupled with genetic analysis will play an important role in documenting whether these events are rare or represent a real shift in large whale movements in the world’s oceans.
Conclusions
Mitochondrial DNA sequence analysis of the unidentified baleen whale that stranded in Louisiana indicate this animal was an Antarctic minke whale, Balaenoptera bonaerensis. Sequences from four nuclear DNA loci corroborate this finding and also indicate that the whale was not a hybrid between an Antarctic minke whale and common minke whale nor a hybrid between Antarctic minke whale and Gulf of Mexico Bryde’s whale. This is the first record for this Southern Hemisphere species in the Gulf of Mexico and, in fact, in the western North Atlantic. It is only the 6th documented record (inclusive of 2 records of minke whale hybrids) for the species in the Northern Hemisphere.
Methods
An unidentified dead baleen whale, 7.7 m in length, was observed by a commercial fisherman on February 5, 2013 floating between South Point and Lake Point in Marsh Island Refuge, Iberia Parish, Louisiana, USA (29.55°, −91.72°; Fig. 1). On the evening of February 10, 2013, members of the Louisiana Department of Wildlife and Fisheries (LDWF), Office of Fisheries were made aware of the stranding. After obtaining additional location information from the original observer, staff were able to access the dead stranded whale and collect samples and photos on February 12, 2013. The animal was assigned the field number CMB-20130212-LA001 and skin for genetic analysis was collected and stored in 20 % DMSO saturated with NaCl. The skull was collected on June 12, 2013. Genomic DNA was extracted from the skin using a Qiagen DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s protocol for extracting DNA from animal tissue. The quantity of extracted DNA was determined using a DyNA Quant 200 fluorometer (GE Healthcare). The sex of the animal was determined as described in Rosel & Wilcox (2014).
The 5’ end of the mitochondrial control region was amplified and sequenced using primers L15824 Rosel et al. 1999) and H16498 (Rosel et al. 1994). The PCR profile included 95 °C for 30 s followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s with a final 7 min 72 °C extension. We used 25 ng of DNA in 25 μl reactions with 20 mM Tris–HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 150 μM dNTPs, 1.0 U Taq DNA Polymerase (Invitrogen), and 0.3 μM of each primer. The resultant PCR product was purified using low melting point gel extraction followed by agarase digestion and the purified product was sequenced commercially (Eurofins MWG Operon) in both directions on a 3730xl Genetic Analyzer (Applied Biosystems). We also amplified and sequenced the 5’ end of the mitochondrial cytochrome b (cytb) gene using the primers L14724 (Palumbi et al. 1991) and H15387 (Rosel et al. 1994). The PCR reagents were as described above for the control region amplification with the addition of 0.08 mg/ml bovine serum albumin. The cycling profile was also as above with the exception that 35 cycles were used and the annealing temperature was 45 °C. The cytb product was purified using Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase (Thermo Scientific) and also commercially sequenced in both directions. All resultant sequences were edited independently using Sequencher 4.5 (Genecodes) and a consensus of the forward and reverse sequences was then created for each gene.
Resultant consensus sequences for the control region and the cytb gene were analyzed two ways to identify the species. Phylogenetic analysis using MrBayes 3.2.3 (Ronquist et al. 2012) was performed on the CIPRES Science Gateway (Miller et al. 2010) to infer the species identity by examining with which baleen whale clade the sequences from the stranded animal grouped. Control region sequences (344 bp, truncated from original 442 bp due to shorter sequences available in GenBank) from all 5 species in the genus Balaenoptera, North Atlantic right whale, Eubalaena glacialis, and humpback whale, Megaptera novaeangliae, were obtained from GenBank and aligned by eye along with the sequence from the unknown stranded animal (Additional file 1: Table S2). jModeltest 2.1.7 (Darriba et al. 2012; Guindon & Gascuel 2003) and the Bayesian information criterion (BIC) were used to determine the best model given the alignments (HKY + G model). The number of substitution sites was set to 2 with gamma-shaped distribution of rates across sites. Bayesian searches involved 4 chains, 2 runs, and 5 000 000 generations using default priors in MrBayes. Burn-in was set to 25 %. Convergence of the runs was determined by examining the average standard deviation of split frequencies and using Tracer 1.6 Rambaut et al. 2014). Second, we applied DNA Surveillance (Ross et al. 2003) to both the control region (442 bp) and cytb (645 bp) sequences obtained from the stranded animal, again applying a phylogenetic approach to species identification. This program uses only vouchered cetacean sequences in its reference database. We performed an ‘Advanced Cluster’ analysis using the Mysticetes Vs4.3 database and 500 bootstrap replicates.
Nuclear sequence data from four loci – ameloblastin (AMBN exon 13), catalase (CAT), int-1 mammary oncogene (INT) and alpha-lactalbumin (LALBA) – were used for genetic comparison of the unidentified stranded animal to sequences of three baleen whale species, the Antarctic minke whale, common minke whale and Bryde’s whale, to investigate whether the stranded whale might be a hybrid between common and Antarctic minke whales as previously described by Glover et al. (2013). These four nuclear loci were chosen based on variability identified by examination of GenBank sequences of balaenopterid species. To further increase the number of individuals sequenced at these loci, we generated, using available tissue samples, common minke whale and Gulf of Mexico Bryde’s whale sequences. Genomic DNA was extracted from the tissue of 13 common minke whale samples from the western North Atlantic using a Qiagen DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s protocol or by standard proteinase K digestion and phenol-chloroform extraction (Rosel & Block 1996). DNA for each nuclear locus was then amplified using previously published primers (Additional file 1: Table S3) for the unidentified baleen whale, the common minke whale samples and 4 Bryde’s whale samples from the GOMx (Rosel & Wilcox 2014). Amplifications were completed as for the mtDNA genes above with the addition of 0.08 mg/ml of bovine serum albumin. The CAT locus was amplified using a touchdown PCR profile as described in Aitken et al. (2004). AMBN exon 13, INT and LALBA were amplified using an initial denaturation step of 94 °C for 2 min, followed by 35 cycles of 94 °C for 45 s, 50 °C for 45 s, and 72 °C for 45 s with a final extension of 72 °C for 7 min. PCR products were purified as described above and sequenced using the Applied Biosystems BigDye Terminator v1.1 cycle sequencing kit. Sequencing products were then purified with Sephadex spin columns before running on an ABI 3130xl Genetic Analyzer. Forward and reverse reads were edited independently using Sequencher 5.2.3 (Genecodes) and a consensus sequence created for each, with the exception of CAT for the unknown whale where a 6 bp deletion in one allele precluded obtaining the full sequence. Additional sequences for the four nuclear loci were obtained from GenBank for the three baleen whale species as available (Additional file 1: Table S2). Sequences were aligned manually for each locus, truncated to the shortest sequence length and identical sequences were identified using MacClade 4.0.8 (Maddison & Maddison 2000). The unique sequences were then examined for variable sites between the unknown sample and the three balaenopterid species (Fig. 4).
Variable sites within each nuclear locus for comparison of the unknown stranded whale to three Balaenoptera species (Bbon = B. bonaerensis, Bacu = B. acutorostrata and Bede = B. edeni from the Gulf of Mexico). Length of alignments for each gene is provided below gene name. N = number of samples with identical sequence. IUB codes used for ambiguous positions, dots indicate identity with top reference sequence, dashes represent gaps. There were no INT sequences available in GenBank for B. bonaerensis. The final column tallies the number of fixed differences and indels (indels counted as events, so a 21 bp indel was counted as a single event) between the unknown baleen whale and other species, but note for some species-loci combinations only a single sequence was available for comparison. Additional file 1: Table S2 provides GenBank accession numbers for sequences taken from GenBank
References
Aitken N, Smith S, Schwarz C, Morin PA. Single nucleotide polymorphism (SNP) discovery in mammals: a targeted-gene approach. Mol Ecol. 2004;13:1423–31.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Deméré TA. Family Balaenopteridae. In: Wilson DE, Mittermeier RA, editors. Handbook of the Mammals of the World. Vol. 4. Sea Mammals. Barcelona: Lynx Edicions; 2014. p. 242–99.
Glover KA, Kanda N, Haug T, Pastene LA, Øien N, Goto M, et al. Migration of Antarctic minke whales to the Arctic. PLoS ONE. 2010;5:e15197.
Glover KA, Kanda N, Haug T, Pastene LA, Øien N, Seliussen BB, et al. Hybrids between common and Antarctic minke whales are fertile and can back-cross. BMC Genet. 2013;14:25.
Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704.
Jackson J, Baker C, Vant M, Steel D, Medrano-Gonzalez L, Palumbi S. Big and slow: phylogenetic estimates of molecular evolution in baleen whales (Suborder Mysticeti). Mol Biol Evol. 2009;26(11):2427–40.
Jefferson TA, Schiro AJ. Distribution of cetaceans in the offshore Gulf of Mexico. Mammal Rev. 1997;27:27–50.
Maddison DR, Maddison WP. MacClade 4: Analysis of phylogeny and character evolution. Version 4.0. Sunderland, MA: Sinauer Associates; 2000.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA; 14 Nov. 2010. pp 1–8.
Mitchell E. Winter records of the minke whale (Balaenoptera acutorostrata Lacepede 1804) in the southern North Atlantic. Rep int Whal Commn. 1991;41:455–7.
Mullin K, Hoggard W, Roden C, Lohoefener R, Rogers C, Taggart B. Cetaceans on the upper continental slope in the north-central Gulf of Mexico. New Orleans, LA: Gulf of Mexico OCS Regional Office; 1991. Contract No.: OCS Study/MMS 91–0027.
Mullin KD, Fulling GL. Abundance of cetaceans in the oceanic northern Gulf of Mexico, 1996–2001. Mar Mamm Sci. 2004;20:787–807.
Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The simple fool’s guide to PCR. Honolulu: University of Hawaii; 1991.
Perrin WF, Brownell Jr RL. Minke whales. In: Perrin WF, Würsig B, Thewissen JG, editors. Encyclopedia of Marine Mammals. 2nd ed. Boston, MA: Academic; 2009. p. 733–5.
Rambaut A, Suchard MA, Xie D & Drummond AJ (2014) Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer 2007.
Rice DW. Marine Mammals of the World. Systematics and Distibution. Lawrence, Kansas USA: Society for Marine Mammalogy (Allen Press); 1998.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Rosel PE, Block BA. Mitochondrial control region variability and global population structure in the swordfish, Xiphias gladius. Mar Biol. 1996;125:11–22.
Rosel P, Wilcox L. Genetic evidence reveals a unique lineage of Bryde’s whales in the northern Gulf of Mexico. Endang Species Res. 2014;25:19–34.
Rosel PE, Dizon AE, Heyning JE. Genetic analysis of sympatric morphotypes of common dolphins (genus Delphinus). Mar Biol. 1994;119:159–67.
Rosel PE, Tiedemann R, Walton M. Genetic evidence for limited trans-Atlantic movements of the harbor porpoise. Phocoena phocoena. Mar Biol. 1999;133:583–91.
Ross HA, Lento GM, Dalebout ML, Goode M, Ewing G, McLaren P, et al. DNA Surveillance: Web-based molecular identification of whales, dolphins, and porpoises. J Hered. 2003;94:111–4.
Segniagbeto GH, Van Waerebeek K, Bowessidjaou JE, Ketoh K, Kpatcha TK, Okoumassou K, et al. Annotated checklist and fisheries interactions of cetaceans in Togo, with evidence of Antarctic minke whale in the Gulf of Guinea. Integr Zool. 2014;9:1–13.
Acknowledgements
We thank Tom Deméré (San Diego Natural History Museum) and Robert Pitman (NMFS SWFSC) for providing expert opinion on the color pattern, skull morphology and species identification of the stranded whale. We also thank NMFS SEFSC staff who coordinated the stranding response and sample collection and the Louisiana stranding network, particularly Damian Bourque and Paul Cook of the Louisiana Department of Wildlife and Fisheries, who responded to the stranding and Brodie Meche who helped with measurement of the skull. A portion of this work was funded by NOAA and BP as part of the Deepwater Horizon Natural Resource Damage Assessment. Stranding response for this animal was funded by the John H. Prescott Marine Mammal Rescue Assistance Grant Program.
Authors’ contributions
PER and LAW designed the study and LAW performed the genetic laboratory work. MCT and CM responded to the reported carcass and carried out field data collection and sample collection. LAW and PER analyzed the data and PER drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Supplementary Tables S1–S3. (PDF 93 kb)
Rights and permissions
Open Access The article is a work of the United States Government; Title 17 U.S.C 105 provides that copyright protection is not available for any work of the United States government in the United States. Additionally, this is an open access article distributed under the terms of the Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0), which permits worldwide unrestricted use, distribution, and reproduction in any medium for any lawful purpose.
About this article
Cite this article
Rosel, P.E., Wilcox, L.A., Monteiro, C. et al. First record of Antarctic minke whale, Balaenoptera bonaerensis, in the northern Gulf of Mexico. Mar Biodivers Rec 9, 63 (2016). https://doi.org/10.1186/s41200-016-0058-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41200-016-0058-x