Abstract
Background
Alien fish species are increasing in the Mediterranean Sea, urging closer monitoring of coastal habitats and biodiversity towards achieving timely assessment and management. Among the alien fish species found in Maltese waters there are members of the Abudefduf genus.
Methods
Research work undertaken with the cooperation of fishermen allows for additional monitoring effort to record the presence of alien species and their possible impacts. One such alien fish specimen collected from Maltese waters in January 2014 was studied through morphological, meristic and genetic analyses. Mitochondrial DNA barcoding analyses were undertaken to confirm the species' identity given that the specimen found belonged to the genus Abudefduf which is composed of a number of congeners that have similar morphological characters.
Results and conclusion
This research led to the identification of the first record of the African Sergeant, Abudefduf hoefleri (Steindachner 1881) (Perciformes: Pomacentridae), in the Mediterranean Sea. This alien tropical species is native of the East Atlantic and may aggressively compete with native Mediterranean reef species for territories and resources.
Similar content being viewed by others
Background
The Pomacentridae is a highly diverse family of reef fishes that primarily inhabit tropical and temperate shallow waters around the world (Allen & Woods 1980; Allen 1991; Bessa et al., 2007; Cooper et al., 2009; Feitosa et al., 2012; Litsios et al. 2012; Froese and Pauly 2013). Till a few years ago, the only representative of this Family in the Mediterranean Sea was the native Chromis chromis (Linnaeus 1758) which is distributed from the eastern Atlantic along the coast of Portugal to the Gulf of Guinea and throughout the Mediterranean Sea (Dulčić 2005; Froese & Pauly 2013). However, in the past two decades, alien species have been on the increase in the Mediterranean Sea (Coll et al., 2010; Golani et al., 2014), including species that are not natural migrants from neighbouring waters (Vacchi et al., 2010; Lipej et al., 2014; Vella et al., 2015 and 2015). Recent records of non-native Pomacentridae members in the Mediterranean include Chrysiptera cyanea (Quoy and Gaimard 1825) that was collected from the Gulf of Trieste, North Adriatic Sea (Lipej et al., 2014) and Stegastes variabilis (Castelnau 1855) that was recently caught from Maltese waters (Vella 2014a and 2014b; Vella et al., 2015a), in the central Mediterranean Sea.
The genus Abudefduf (Linnaeus, 1758) was first recorded in the Mediterranean in 1957 (Tardent, 1959), with several more recent records of A. vaigiensis (Goren & Galil, 1998; Vacchi & Chiantore, 2000; Golani et al., 2014; Vella 2014a and 2014b), and A. saxatilis (Azzurro et al., 2013; Vella 2014a and 2014b; Deidun & Castriota 2014; Tsadok et al., 2015) at various coastal locations around the Mediterranean. However, there have been no records of the African sergeant, A. hoefleri in this region. This fish is native to the tropical Eastern Atlantic region, ranging from Senegal to Benin, including Ilheu das Rolas, Sao Tome and Cape Verde (Loris & Rucabado 1990; Edwards et al., 2001; Cooper et al., 2009; Cowman & Bellwood, 2013), with its most northern records being the Canary Islands (Cooper et al., 2009; Triay-Portella et al., 2015).
Results and discussion
Morphometrics
The specimen caught weighed 94.98 g and had a total body length of 174.0 mm. This length is close to the maximum size recorded for this species (Froese & Pauly, 2013). Its appearance, morphology and meristics presented in Tables 1 and 2 and Figs. 1 and 2 matched the descriptions of A. hoefleri given by Allen (1991), Edwards et al. (2001), and Froese & Pauly (2013). This specimen had XIII + 14 dorsal finrays, II + 13 anal finrays, I + 5 ventral fin rays and 18 pectoral finrays. The body had a dark bluish colouration with four darker blue strips which were mostly pronounced just after being caught (Fig. 1).
Genetic analyses
A total of 3992 bp were sequenced from the A. hoefleri specimen's mtDNA. The sequence lengths obtained were 606 bp, 833 bp, 458 bp, 829 bp and 1266 bp for COI, ATPase, nd3, cytb and 12S-16S respectively, and each was run via BLASTn to identify sequence matches. All the genes confirmed the genus with high identity matches. At species level, the corresponding sequences available in GenBank are limited to one specimen collected from the Canary Islands, studied by Cooper et al. (2009). Therefore, it was not possible to compare all the currently studied genes with other species specific genetic data. Both the current study and that by Cooper et al. (2009) analysed part of the 12S rRNA gene and nd3. When the latter genes were compared against each other, 352 bp of the 12S rRNA gene gave 100 % identity match while 408 bp of nd3 gave a 99.8 % identity match with FJ616290 and FJ616506 respectively Cooper et al. (2009), thus confirming the species identity. Additionally, phylogenetic analyses of the genus have placed the currently studied A. hoefleri specimen on a distinct branch, with the closest congeners being A. troschelii and A. saxatilis (Fig. 3).
Discussion
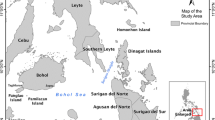
Unlike most of the alien fish species noted in the Mediterranean which have entered through the Suez Canal (Occhipinti-Ambrogi et al., 2011; Kalogirou et al. 2012; Golani et al., 2010; Golani et al., 2014; Vella et al., 2015b), this new alien species has probably entered through the Strait of Gibraltar, given that its typical range is of tropical eastern Atlantic origin (Loris & Rucabado 1990; Edwards et al., 2001; Cowman & Bellwood, 2013). The lack of records of A. hoefleri in other regions of the Mediterranean Sea might indicate that this species is not a natural migrant but has been introduced through shipping activity, especially given that the occurrence of this species has been noted to coincide with the presence of port activities and oil platforms in the Canary Islands (Triay-Portella et al., 2015). At Mediterranean level, shipping activity is one of the main vectors introducing alien marine species in the region (Galil, 2006; Katsanevakis et al., 2014). The record presented here occurred in a location very close to harbours, transhipment hubs and off-shore bunkering sites in the Malta. Moreover, the specimen was caught from the semi-enclosed embayment of the Ħofra ż-Żgħira (Fig. 4), an area that receives thermal effluent discharges from a nearby power station. On exit, this effluent has a temperature higher than the ambient temperature, causing the water at the Ħofra ż-Żgħira to have temperatures up to 8 °C warmer than surrounding ambient temperatures (Enemalta, 2011). The warmer environmental conditions at this site may have helped this tropical species survive. Nonetheless one cannot exclude the possibility that this species might be a very recent natural migrant which has not been yet recorded in other stepping stone locations and is expanding its natural home range, possibly colonizing regions that are undergoing gradual warming. The latter being reported as facilitating immigration and the survival of exotic species in the Mediterranean Sea (Occhipinti-Ambrogi & Galil, 2010).
Conclusion
Ongoing monitoring may verify the spread or loss of this non-indigenous species in these waters. The already at risk Mediterranean fish species (Coll et al. 2010) are being faced with increasing threats from alien species (Coll et al., 2015). As a member of the genus Abudefduf, this new alien can be aggressively competitive towards other fishes, therefore like other alien species its occurrence can have detrimental effects to coastal fishes communities with potential negative repercussions on marine conservation of local species.
Methods
On the 4th of January 2014 a specimen of A. hoefleri (Figs. 1 and 2) was caught in shallow coastal waters close to Delimara, Malta [GPS: 35.835260 N, 14.562635E] (Fig. 4) during sport-fishing being monitored by researchers from the Conservation Biology Research Group of the University of Malta. Upon capture the whole specimen was kept for scientific analyses. The diagnostic features that were used in the morphological identification of the specimen were analysed according to Allen (1991), Edwards et al. (2001), and Froese & Pauly (2013). All length measurements were taken to the nearest 0.1 mm using electronic calipers and mass was recorded to the nearest 0.01 g.
Tissue samples were collected from this specimen and preserved in 95 % ethanol. The specimen was subsequently deposited in the ichthyological collection of the Conservation Biology Research Group laboratory at the University of Malta with reference code number CBRG/F.140104/AH001. For genetic analyses, the tissue was digested with Proteinase K and the total DNA was extracted using AccuPrep® Genomic DNA Extraction Kit (Bioneer). PCR amplifications were carried out for: cytochrome c oxidase I gene (COI) using FishF2 and FishR2 primers (Ward et al., 2005); ATP synthase subunit 6 and its flanking protein coding genes (ATPase) using L8331 (Meyer 1993) and H9236 primers (Quenouille et al., 2004); NADH dehydrogenase subunit 3 and its flanking tRNA genes (nd3) using nd3-F270 and nd3-R750 primers (Cooper et al., 2009); cytochrome b (cytb) using GluDG-L14724 and CB3H-15560 primers (Martin and Palumbi 1993); and 12S to 16S ribosomal RNA genes (12S-16S) using 12SA and 16SA primers (Palumbi 1996). The amplification for each primer set was performed using an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 52 °C for 45 s and extension at 72 °C for 40 s and a final extension at 72 °C for 10 min, except 12S-16S where an annealing temperature of 48 °C was used. The PCR products were purified and sequenced via ABI3730XL sequencer using both the forward and reverse primers. The sequences, at both nucleotide and amino acid level, were analyzed using Geneious v6.1.6 (http://www.geneious.com, Kearse et al., 2012). The sequences obtained were deposited in GenBank accession numbers KT374287-91 for COI, ATPase, nd3, cytb and 12S-16S respectively. These sequences were compared to other sequences available in genomic databases using BLASTn.
Various COI gene sequences for different Abudefduf species available in GenBank and BOLD, allowed for phylogenetic analyses of this specimen. The sequences were aligned using Geneious v6.1.6 (http://www.geneious.com, Kearse et al., 2012). A 549 bp sequence representing the smallest homologous COI sequence was selected and genetic divergences were calculated using the Kimura 2-parameter distance model (Kimura 1980), while the phylogenetic tree was constructed using 1500 bootstraps with Maximum Likelihood utilizing MEGA v5.2.1 Tamura et al. (2011).
Availability of supporting data
The genetic sequence data supporting the results of this article are available in GenBank, as accession numbers KT374287-91 for COI, ATPase, nd3, cytb and 12S-16S respectively.
References
Allen GR. Damselfishes of the world. Melle, Germany: Mergus Publishers; 1991. 271.
Allen GR, Woods LP. A review of the damselfish genus Stegastes from the Eastern Pacific with description of a new species. Records of the Western Australian Museum. 1980;8:171–98.
Azzurro E, Broglio E, Maynou F, Bariche M. Citizen science detects the undetected: the case of Abudefduf saxatilis from the Mediterranean Sea. Management of Biological Invasions. 2013;4:167–70.
Bessa E, Dias JF, de Souza AM. Rare data on a rocky shore fish reproductive biology: sex ratio, length of first maturation and spawning period of Abudefduf saxatilis (Linnaeus, 1758) with notes on Stegastes variabilis spawning period (Perciformes: Pomacentridae) in São Paulo, Brazil. Braz J Oceanogr. 2007;55:199–206.
Coll M, Piroddi C, Steenbeek J, Kaschner K, Frida Ben Rais F, Aguzzi J, Ballesteros E, Bianchi CN, Corbera J, Dailianis T, Danovaro R, Estrada M, Froglia C, Galil BS, Gasol JM, Gertwagen R, Gil J, Guilhaumon F, Kesner-Reyes K, Kitsos MS, Koukouras A, Lampadariou N, Laxamana E, Lopez-Fede la Cuadra CM, Lotze HK, Martin D, Mouillot D, Oro D, Raicevich S, Rius-Barile J, Saiz-Salinas JI, San Vicente C, Somot S, Templado J, Turon X, Vafidis D, Villanueva R, Voultsiadou E. The biodiversity of the Mediterranean Sea: estimates, patterns and threats. PLoS ONE. 2010;5:e11842.
Coll M, Steenbeek J, Ben Rais Lasram F, Mouillot D, Cury P. 'Low-hanging fruit' for conservation of marine vertebrate species at risk in the Mediterranean Sea. Global Ecology and Biogeography. 2015;22:226–239.
Cooper W, Smith LL, Westneat MW. Exploring the radiation of a diverse reef fish family: phylogenetics of the damselfishes (Pomacentridae), with new classifications based on molecular analyses of all genera. Mol Phylogenet Evol. 2009;52:1–16.
Cowman PF, Bellwood DR. Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proc R Soc Lond B Biol Sci. 2013;280:20131541.
Deidun A, Castriota L. First record of Abdudefduf cfr saxatilis Linnaeus, 1758 (Perciformes:Pomacentridae) from the Maltese Islands (Central Mediterranean). BioInvasions Records. 2014;3:53–6.
Dulčić J. Biometric properties of damselfish, Chromis chromis (Osteichthyes: Pomacentridae) from the middle Adriatic. Acta Adriat. 2005;46:91–8.
Edwards AJ, Gill AC, Abohweyere P.O. A Revision of Irvine's Marine Fishes of Tropical West Africa. Darwin Initiative Report 2, Ref. 162/7/451. 2001. p. 157. http://www.ncl.ac.uk/tcmweb/tmr/Irvines_Marine_Fishes.PDF. Accessed 10th April 2013.
Enemalta. Prediction of the Spread and Dilution of Cooling Water from Delimara Power Station; SLR global environmental solutions Report Ref. 403.0082.00022; 2011. 41. URL http://www.mepa.org.mt/lpgdocuments/thermal%20Model.pdf. Accessed 20th August 2015
Feitosa JLL, Concentino AM, Teixeira SF, Ferreira BP. Food resource use by two territorial damselfish (Pomacentridae: Stegastes) on South-Western Atlantic algal-dominated reefs. J Sea Res. 2012;70:42–9.
Froese R. and Pauly D. FishBase. 2013. URL http://www.fishbase.org. Accessed 10th April 2013
Galil BS. Shipwrecked – Shipping impacts on the biota of the Mediterranean Sea. Chapter 3. In: Davenport J, Davenport JL, editors. The Ecology of Transportation: Managing Mobility for the Environment. The Netherlands: Springer publishers; 2006. p. 392.
Golani D. Colonization of the Mediterranean by Red Sea fishes via the Suez Canal- Lessepsian migration. In: Golani D, Appelbaum-Golani B, editors. Fish Invasions of the Mediterranean Sea: Change and Renewal. Sofia-Moscow: Pensoft Publishers; 2010. p. 145–88.
Golani D, Orsi-Relini L, Massutí E, Quignard JP. CIESM - Atlas of Exotic Fishes - List [WWW Document]. 2014. URL http://www.ciesm.org/atlas/appendix1.html. Accessed 20th August 2015.
Goren M, Galil BS. First record of the Indo-Pacific, coral-reef fish Abudefduf vaigiensis (Quay & Gaimad, 1825) in the Levant. Isr J Zool. 1998;44:57–9.
Kalogirou S, Azzurro E, Bariche M. The Ongoing Shift of Mediterranean Coastal Fish Assemblages and the Spread of Non-Indigenous Species, Chapter 11. In A.G. Lameed (ed) Biodiversity Enrichment in a Diverse World, InTech;2012. 263–280. http://www.intechopen.com/books/biodiversity-enrichment-in-a-diverse-world/the-ongoing-shift-of-mediterranean-coastal-fish-assemblages-and-the-spread-of-non-indigenous-species. Accessed 20th August 2015
Katsanevakis S, Coll M, Piroddi C, Steenbeek J, Ben Rais Lasram F, Zenetos A, Cardoso AC. Invading the Mediterranean Sea: biodiversity patterns shaped by human activities. Frontiers in Marine Science. 2014. doi:10.3389/fmars.2014.00032.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Lipej L, Mavri B, Dulcic J. First record of Chrysiptera cyanea (Quoy and Gaimard, 1825) (Perciformes: Pomacentridae) in the Mediterranean Sea. J Appl Ichthyol. 2014;30(5):1–3.
Litsios G, Pellissier L, Forest F, Lexer C, Pearman PB, Zimmermann NE, and Salamin N. Trophic specialization influences the rate of environmental niche evolution in damselfishes (Pomacentridae). Proc R Soc B Biol Sci. 2012;279:3662–9.
Loris D, Rucabado J. Pomacentridae. In: Quéro JC, Hureau JC, Karrer C, Post A, Saldanha L, editors. Check-list of the fishes of the eastern tropical Atlantic (CLOFETA). Volume 2. Lisbon: JNICT; 1990. p. 842–50. SEI, Paris; and UNESCO, Paris.
Martin AP, Palumbi SR. Protein evolution in different cellular environments: cytochrome b in sharks and mammals. Mol Biol Evol. 1993;10:873–91.
Meyer A., 1993. Evolution of mitochondrial DNA in fishes. In: Hochachka PW, Mommsen TP (Eds.), Biology Frontiers, Biochemistry and Molecular Biology of Fishes, vol. 2. Elsevier Science, Amsterdam, pp. 1–38.
Occhipinti-Ambrogi A, Galil B. Marine alien species as an aspect of global change. Adv Oceanogr Limnol. 2010;1:199–218.
Occhipinti-Ambrogi A, Marchini A, Cantone G, Castelli A, Chimenz C, Cormaci M, Froglia C, Furnari G, Gambi MC, Giaccone G, Giangrande A, Gravili C, Mastrototaro F, Mazziotti C, Orsi-Relini L, Piraino S. Alien species along the Italian coasts: an overview. Biol Invasions. 2011;13:215–37.
Palumbi SR. Nucleic Acids II: The Polymerase Chain Reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Massachusetts: Sinauer Associates; 1996. p. 205–47.
Quenouille B, Bermingham E, Planes S. Molecular systematics of the damselfishes (Teleostei: Pomacentridae): Bayesian phylogenetic analyses of mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 2004;31:66–88.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Tardent P. Capture d'un Abudefduf saxatilis vaigiensis Q. und G. (Pisces, Pomacentridae) dans le Golfe de Naples. Rev Suisse Zool. 1959;66:347–51.
Triay-Portella R, Pajuelo JG, Manent P, Espino F, Ruiz-Diaz R, Lorenzo JM, Gonzalez JA. New records of non-indigenous fishes (Perciformes and Tetraodontiformes) from the Canary Islands (north-eastern Atlantic). Cybium. 2015;39(3):163–74.
Tsadok R, Rubin-Blum M, Shemesh E, Tchernov D. On the occurrence and identification of Abudefduf saxatilis (Linnaeus, 1758) in the easternmost Mediterranean Sea. Aquat Invasions. 2015;10:101–5.
Vacchi M, Chiantore MC. Abudefduf vaigiensis (Quoy & Gaimard, 1825): a tropical damselfish in Mediterranean Sea. Biologia Marina Mediterranea. 2000;7:841–3.
Vacchi M, Psomadakis PN, Repetto N, Würtz M. First record of the dog snapper Lutjanus jocu in the Mediterranean Sea. J Fish Biol. 2010;76:723–8.
Vella A. Conservation research reports new alien species and declining local species in our sea. 2014a. The Maritime Directory Website news: http://www.maritimedirectory.com.mt/newsread.asp?l=e&ID=3037 issued on the 16th of January 2014. Accessed 10th May 2015.
Vella A. Conservation research reports new alien species and declining local species in our sea. 2014b. The University of Malta Website news: http://www.um.edu.mt/newsoncampus/researchinitiatives/archive/newaliendeclininglocalspecies. Accessed 10th May 2015.
Vella A, Agius Darmanin S, Vella N. Morphological and genetic barcoding study confirming the first Stegastes variabilis (Castelnau, 1855) report in the Mediterranean Sea. Marine Mediterranean Science. 2015a;16:609–12.
Vella A, Vella N, Agius Darmanin S. First record of Lutjanus fulviflamma (Osteichthyes: Lutjanidae) in the Mediterranean Sea. J Black Sea/Mediterranean Environment. 2015b;21:307–15.
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci. 2005;360:1847–57.
Acknowledgements
Thanks are due to sport fishermen, in particular to Mr. S. Zammit who has cooperated with this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AV has contributed to all aspects of the research work presented here including the conception and design of the molecular genetics research, analyses and interpretation of both genetic and morphological data and was involved in finalizing the manuscript. NV contributed to molecular genetic research, analyses and interpretation. SAD contributed to the morphological research work of the specimen collected. All three authors were involved in the drafting of the manuscript and in giving approval for the version for publication.
Authors’ information
All three authors are researchers of the Conservation Biology Research Group, Department of Biology, University of Malta.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Vella, A., Vella, N. & Agius Darmanin, S. The first record of the African Sergeant, Abudefduf hoefleri (Perciformes: Pomacentridae), in the Mediterranean Sea. Mar Biodivers Rec 9, 15 (2016). https://doi.org/10.1186/s41200-016-0008-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41200-016-0008-7