Abstract
Purpose
This study aims to reveal the relationship between AMIGO2 and proliferation, migration and tumorigenicity of bladder cancer, and explore the potential molecular mechanisms.
Methods
The expression level of AMIGO2 is measured by qRT-PCR and immunohistochemistry (IHC). Stable AMIGO2 knockdown cell lines T24 and 5637 were established by lentivirus transfection. Cell Counting Kit (CCK-8 assay) was produced to determine cell proliferation, flow cytometry analysis was utilized to detect cell cycle, and wound healing assay was proceeded to test migration ability of bladder cancer cells. Xenograft mouse model was established for investigating the effect of AMIGO2 on tumor formation in vivo. The RNA Sequencing technology was applied to explore the underlying mechanisms. The expression level of PPAR-γ was measured by Western Blot.
Results
AMIGO2 was upregulated in bladder cancer cells and tissues. Inhibited expression of AMIGO2 suppresses cell proliferation and migration. Low AMIGO2 expression inhibited tumorigenicity of 5637 in nude mice. According to RNA-Seq and bioinformatics analysis, 917 DEGs were identified. The DEGs were mainly enriched in cell–cell adhesion, peroxisome proliferators-activated receptors (PPARs) signaling pathway and some other pathways. PPAR-γ is highly expressed in bladder cancer cell lines T24 and 5637, but when AMIGO2 is knocked down in T24 and 5637, the expression level of PPAR-γ is also decreased, and overexpression of PPAR-γ could reverse the suppression effect of cell proliferation and migration caused by the inhibition of AMIGO2.
Conclusion
AMIGO2 is overexpressed in bladder cancer cells and tissues. Knockdown of AMIGO2 suppresses bladder cancer cell proliferation and migration. These processes might be regulated by PPAR-γ signaling pathway.
Similar content being viewed by others
Introduction
Bladder cancer (BCa) is the 6th most common cancer in male (4.4% of all cancers) and the 10th most common cancer in both sex (3.0% of all) [1]. Urothelial carcinoma of the bladder is a prevalent cancer type that causes approximately 150,000 deaths globally each year [2]. Thus, exploring more treatment options is of utmost importance. Novel approaches are currently surfacing, including cancer vaccines, targeted therapeutic regimens, immune modulation strategies, nanoparticle-mediated therapies, cytotoxic agents, hyperthermia, and gene therapy applications [3]. Among all the therapies, gene therapy stands out as a particularly promising contender. Consequently, the exploration of new targets is important, and in-depth studies of the potential molecular mechanisms underlying the biological behavior of bladder cancer cells would aid the identification of reliable molecular markers for bladder cancer patients.
The adhesion molecule with Ig-like domain 2 (AMIGO2) located in the 12th chromosome, precisely within the chromosomal region q13.11, spanning a length of 3175 bp. The protein encoded by AMIGO2 exhibits amphoteric properties and is characterized by an extracellular domain featuring 6 leucine repeat sequences (LRR), along with an Ig-like domain [4]. AMIGO2 plays a crucial role in various cellular processes, notably including tumor growth, cell adhesion and migration through collagen of gastric cancer cells [5]. In addition, AMIGO2 has been identified as a critical determinant in the promotion of melanoma cell proliferation and survival. This effect appears to be facilitated by the acquisition of active cis-acting DNA regulatory elements that are notably absent in Normal Human Melanocytes (NHMs) [6].
The peroxisome proliferator-activated receptor-gamma (PPAR-γ) represents a prominent constituent within the hormone-responsive nuclear receptor superfamily. As is reported before, PPAR-γ induces antiproliferative and antiangiogenic pathways across various tissue types, thus making it a promising target for downregulation of carcinogenesis [7]. In neuroblastoma, PPAR-γ plays a tumor-promoting role. In contrast, in breast cancer cells, PPAR-γ inhibits inflammatory response, and it functions as a tumor suppressor. Therefore, the research suggests that the ability of PPAR-γ to promote or suppress tumor formation is linked to cell type-specific differences in target genes [8]. In hepatocellular carcinoma, instances of functional impairment stemming from mutations affecting suppressive regulators of PPAR-γ, such as histone deacetylase 3 (HDAC3) and nuclear hormone corepressor (N-CoR), engender a propensity toward steatosis and aberrant hepatic expansion, ultimately culminating in malignancy in both murine models and human subjects [9, 10]. Therefore, the function of PPAR-γ across different cancer types seems to be controversial [11].
In the present study, we explored the biological functions of AMIGO2 in bladder cancer. Our study illuminated that AMIGO2 promotes the proliferation, migration and tumorigenicity of BCa cells by regulating PPAR-γ pathway. These findings deepened our understanding of the molecular mechanisms underlying the progression of bladder cancer. These results might hold promise for finding potential therapeutic targets of bladder cancer.
Materials and methods
Cell culture and tissue specimen collection
The human uroepithelium cell line SV-HUC-1, human bladder cancer cell lines T24 and 5637 were obtained from Chinese Academy of Science, authenticated by STR profiling and tested for mycoplasma contamination. SV-HUC-1 was cultured in F-12 K medium, and T24 and 5637 were cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, USA), supplemented with 10% fetal bovine serum (Pan Biotech, Germany), penicillin and streptomycin (100 IU/ml). All the cells were incubated at 37℃ under a humidified atmosphere with 5% CO2.
Bladder cancer tissues and their matched adjacent non-tumor bladder tissues (n = 16) were collected from patients who underwent radical cystectomy and received a pathological diagnosis of bladder cancer at the Department of Urology, Lanzhou University Second Hospital. All the samples were promptly harvested post-surgery, fixed in formalin, and subsequently embedded in paraffin. None of the enrolled patients underwent chemotherapy, radiotherapy or adjuvant treatment before surgery. All tissue acquisitions were conducted in accordance with the principles of informed consent, with each patient providing written consent for the utilization of their tissue samples in this research. This project was approved by the Institutional Research Ethics Committee of Lanzhou University Second Hospital (No. 2019A-119).
Tissue Microarray (TMA) construction and image acquisition
The construction of TMAs was executed in accordance with the method outlined previously [12]. Briefly, tissue blocks were meticulously arranged in the desired order. The selection of the appropriate tool diameter, which in our study was set at 2.0 mm, was a critical step. Subsequently, up to 9 TMA blocks were loaded into the apparatus, and distinct identification labels were assigned to each of the 9 blocks. Establish a TMA layout for each recipient block. Following this, the donor blocks were introduced into the tissue microarrayer, and their placement within each row was determined based on their respective case numbers. Each donor block was then attributed with a unique donor ID. Use the punching tool, this instrument punches a hole from the donor block at the selected region. Cores are then transferred from the donor blocks to the recipient one. After the completion of punching holes in the first round, the process moves on to the next round, following the same steps described before. Repeat this process until all the available donor block have been punched. At this point, the production of TMA is finished.
TMA images were viewed and acquired using panoramic scanning microscope (TissueFAXS Viewer 7.1.6245.119, Austria). A total of 16 pairs of cancer tissues and corresponding normal tissues were included in the cohort. One representative image was meticulously chosen for each patient. All images derived from the TMAs were systematically incorporated into the subsequent stages of image processing analysis.
H&E (Hematoxylin–eosin) staining
Routine dewaxing and hydration were performed on the tissue slices. After staining with hematoxylin for 5 min, differentiation was carried out using 1% hydrochloric acid ethanol. Following a 10-min staining with eosin, the slices were cleared in xylene, dehydrated in alcohol, and mounted with neutral resin. Imaging was captured and viewed using panoramic scanning microscope (TissueFAXS Viewer 7.1.6245.119, Austria).
Immunohistochemiscal analysis
An IHC analysis was performed using formalin-fixed, paraffin-embedded tissues as previously described [13]. Briefly, the TMA sections were first deparaffinized in xylene and rehydrated in graded alcohol. For antigen retrieval, tissue slides underwent a process immersion and boiling in a 0.01 M citrate buffer solution for 16 min. Subsequently, these slides were incubated in a humidified chamber with AMIGO2 antibody (1:250, Santa Cruz, USA) overnight at 4℃. Following the primary antibody incubation, the slides were subjected with biotinylated anti-goat IgG secondary antibody for 15 min at room temperature. Sections were then stained with 3, 3`-diaminobenzidine (DAB) and counterstained with hematoxylin (Solarbio, Peking, China). For the evaluation of IHC staining, the samples were scored based on two parameters. The first parameter considered the proportion of positively stained cells, categorized as follows: 0-none, 1—< 25%, 2—25–50%, 3—50–75%, and 4—75–100%. The second parameter assessed staining intensity, with scores assigned as follows: 0—none, 1—weak, 2—medium and 3—strong. To derive a comprehensive score, these two sub-scores were multiplied. Samples achieving total scores in the range of 0–6 were classified as low expression, while 7–12 were considered as high expression. The patients’ information and IHC Scores are listed in Table 1. The clinicopathologic characteristics of 16 cases are documented in Table 2.
Cell transfection
Plasmids carrying AMIGO2, PPAR-γ and negative control shRNA were constructed. Lentivirus was generated in HEK-293 cells. The plasmid was procured from Shanghai Genechem Co., LTD. Lentivirus plasmid constructed with scramble sequence was used as control. The transfection procedure was executed following the manufacturer’s instructions. Puromycin was introduced at a concentration of 1.5 µg/ml for selection. Cell clones that exhibited stable down-regulation of AMIGO2 (shAMIGO2) and those maintained as negative control (shCtrl) were selected and expanded within T24 and 5637. Then, PPAR-γ was stably overexpressed in shAMIGO2 group and shCtrl group in both cell lines, following the same procedure described before.
RNA extraction, reverse transcription and quantitative real-time PCR
Total RNAs were extracted from cultured cells using Trizol Reagent (Takara, Japan) according to the manufacturer’s instructions. Conversion to cDNA was achieved through cDNA PrimeScript™ RT Master Mix (Perfect Real Time) (TAKARA, Japan) in reverse transcription PCR instrument (Bio-Rad Laboratories, USA). qRT–PCR was carried out using the CFX Real-Time PCR System (Bio-Rad Laboratories, USA) in a 15-μl reaction volume containing first-strand cDNA, TB Green™ Premix Ex Taq™ II (Tli RNaseH Pluse). The relative expression was calculated using the 2−ΔΔCt method [14]. The transcription level of GAPDH was used as an internal control. The primers used are listed in Table 3.
Protein extraction and Western Blot
Cells were thawed on ice by ultrasonication in RIPA buffer (Beyotime, China). The resulting homogenates were centrifuged at 4℃, 12000 g, for 15 min, followed by the collection of the supernatants. Protein concentration was measured by BCA Protein Assay Kit (Beyotime, China). Protein isolates were then resolved on SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were blocked with 5% milk, incubated overnight with the primary antibodies: Anti-AMIGO2 (Santa-Cruz, USA); Anti-PPAR-γ (Abcam, UK); β-Actin (Santa-Cruz, USA), and probed with horseradish peroxidase-conjugated secondary antibody: Anti-Mouse IgG (Santa-Cruz, USA). The blots were then detected using Pierce™ ECL Western Blotting Substrate Kit (Thermo, USA).
Flow cytometric cell cycle analysis
Cells (1 × 106) were collected, re-suspended in PBS, fixed in 75% ice-cold ethanol, and incubated in propidium iodide (PI, 50 µg/ml; Sigma, USA) in the dark for 20 min. RNA enzyme (100 µg/ml) was added into the system. Cell cycle analysis was performed using the BD LSRII Flow Cytometry System with FACSDiva software (BD Bioscience, Franklin Lakes, USA). The data were analyzed using the Modfit LT software.
CCK-8
Cells were seeded into 96-well plates (2000 cells/well). Cell proliferation was detected using the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Japan). At the specified time point, CCK-8 solution was added to each well and incubated for 3 h. The absorbance was measured at 450 nm with a microplate meter (Tecan infinite, Switzerland).
Wound-healing assay
3 × 105 T24 or 5637 cells were inoculated into 6-well plates with complete culture medium separately. The confluent monolayer of both cells was scratched with a 1 ml pipette tip. The cells were then washed with PBS three times and kept in the medium (supplemented with 10% FBS) for 72 h. The wound width was recorded at 0 h, 24 h, 48 h and 72 h under a microscope.
Xenograft mouse model
Nude mice (BALB/c-nu, female, 4–6 weeks, 16-18 g) were bred and housed in AAALAC-accredited specific pathogen-free rodent facilities. Mice were accommodated in sterilized and ventilated microisolator cages, receiving ample autoclaved commercial chow and sterile water. The mice were randomly divided into two groups (n = 8) and housed in four cages (4 mice per cage). Cage size: 310 × 230 × 160 mm. The environmental conditions were rigorously controlled. Ambient temperature was maintained within the range of 20–25℃, with humidity levels sustained between 40 and 70%. Lighting conditions followed a 12-h light–dark cycle. The overall living conditions were stable. Tumorigenicity was determined by subcutaneously injection of 5637 cells into the flanks of female nude mice (5 × 106 cells per site). Tumors were measured every 2 days using an electronic skin caliper, capturing the longest width and length. Tumor volumes were calculated employing the formula: V = π/6 × largest diameter × smallest diameter2 [15]. No blinding was done. All mouse experiments were conducted with standard operating procedures approved by the University Committee on the Use and Care of Animals at Lanzhou University Second Hospital (Approval No. D2019-171).
RNA-Seq data analysis
Total RNAs were extracted from both shAMIGO2 and shCtrl samples. We employed RNA-Seq technology (Novogene, China) for the identification of DEGs (Differentially Expressed Genes) [16]. In brief, the raw fastq data generated from RNA-seq were initially subjected to trimming using Trimmomatic (V0.35). Subsequently, the trimmed reads underwent alignment to the human reference genome (NCBI GRCh38) utlizing TopHat (Version 2.0.12) with default parameter settings. The resulting aligned bam files were then processed using Cufflinks (Version 2.2.1) for gene quantification. For an additional layer of analysis, reads were also mapped to ERCC (External RNA Controls Consortium) transcripts and quantified using TopHat (Version 2.0.12) and Cufflinks (Version 2.1.1) with default parameter settings. Genes meeting the threshold of FPKM ≥ 1 (Fragments Per Kilobase of transcript per Million mapped reads) across all samples were included in the subsequent analysis to identify DEGs. We could upload the data from RNA-Seq to a public repository if necessary.
Data analysis and DEGs identification
The sequencing dataset underwent normalization and analysis through the DESeq package (Version 1.10.1). The criteria of FDR < 0.01 (False Discovery Ratio) and |logFC|> 2 (Fold Change) was set as the threshold. Visualization of the dataset was conducted using the R programming software, generating both a volcano plot and a heat map. The most upregulated 100 genes and downregulated 100 genes were chosen for the heat map.
Biological function and pathway enrichment analysis
Through calculating the corresponding topological overlap, genes positively associated with AMIGO2 were found out and subjected to gene ontology (GO) analysis (GOSeq, Release2.12) to determine clusters of DEGs with enriched molecular functions. Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis was performed via the “clusterProfiler” package in R/Bioconductor software to acquire the enriched biological process and KEGG pathway. p < 0.05 and counts ≥ 4 were considered significant.
Module analysis of protein–protein interaction (PPI) network
Search Tool for the Retrieval of Interacting Genes (STRING) database (http://www.string-db.org/) was applied to acquire PPI information for the DEGs. Cytoscape software (3.7.2) was applied to visualize the PPI network. The top DEGs with a high degree of connectivity in the PPI network were selected to discuss their function and effect on bladder cancer.
Kaplan–Meier survival analysis
OS survival data of TCGA samples was downloaded from the Xena database (https://xenabrowser.net/) to study the prognostic significance of hub genes. Kaplan–Meier method was applied for survival analysis with packages “survival” and “survminer”.
Statistical analysis
All statistical analyses were carried out using IBM SPSS Statistics 20 (SPSS Inc., Chicago, IL, USA). The two-tailed Student’s t-test was used to evaluate statistical differences between two groups. The correlation between AMIGO2 expression level and patients’ clinicopathological characteristics were analyzed using Chi-squared test. The survival curve was described utilizing the Kaplan–Meier plot. *p < 0.05; **p < 0.01; and ***p < 0.001 were considered statistically significant.
Results
AMIGO2 is upregulated in bladder cancer cells and tissues
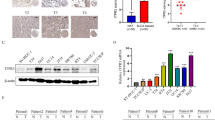
The result of qRT-PCR showed that the AMIGO2 expression level was elevated in bladder cancer cell lines T24 and 5636 compared with SV-HUC-1 (Fig. 1A). The relative mRNA expression of AMIGO2 in bladder cancer tissues was significantly higher than that of their matched adjacent normal tissues (n = 11) (Fig. 1B). The protein expression level of AMIGO2 was also up-regulated in bladder cancer tissues compared with the matched adjacent normal tissues (n = 16) (Fig. 1C, D). HE and IHC staining were applied to identify. However, as shown in Table 2, according to current evidence, the expression of AMIGO2 is not associated with age (P = 0.074), gender (P = 0.064), TNM classification (P = 0.687; P = 0.551 and P = 0.383), pathological grade (P = 0.551) or vascular invasion (P = 0.182).
Expression level of AMIGO2 in bladder cancer cells and tissues. A mRNA expression of AMIGO2 in one normal bladder epithelial cell line SV-HUC-1 and two bladder cancer cell lines: T24 and 5637. B The mRNA expression level of AMIGO2 was examined in cancer tissues (T) and adjacent normal tissues (ANT) (n = 11). C Representative HE and IHC images of AMIGO2 protein expression in cancer tissues and the corresponding adjacent normal tissues (n = 16). All images were captured at 50 × and 100 × magnification. D The violin plots of AMIGO2 protein expression level in all 16 pairs of tissue samples. Data were based on at least three independent experiments, and shown as mean ± SEM (*p < 0.05, ** p < 0.01, *** p < 0.001). BLCA: Bladder Cancer. TMA: Tissue Microarray
Inhibition of AMIGO2 suppresses proliferation and migration in vitro
CCK-8 and wound healing assay showed that suppression of AMIGO2 decreased the proliferation and migration capacity of T24 and 5637 cells in shAMIGO2 group compared with shCtrl group (Fig. 2A, B). Flow cytometry displayed an increase in the percentage of cells in G1/G0 phase and a decrease in the percentage of cells in S phase (Fig. 2C). All these results supported that downregulation of AMIGO2 reduces the proliferation and migration and induces G1 phase cell cycle arrest.
Knockdown of AMIGO2 suppresses proliferation, migration and cell cycle in bladder cancer. A Effects of shAMIGO2 and shCtrl on the proliferation of bladder cancer cell lines T24 and 5637, detected by CCK-8. B Effects of shAMIGO2 and shCtrl on the migration BCa cells, assessed by wound healing assay. C Effects of shAMIGO2 and shCtrl on cell cycle of the indicated cells, analyzed by flow cytometry. Bars represent the mean ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001
Inhibition of AMIGO2 reduces tumorigenicity of bladder cancer cells in vivo
The growth of tumors derived from the shAMIGO2 group was prominently suppressed compared with the shCtrl group after 5637 injection (n = 8), detected for 36 days (Fig. 3A, B). These results indicated that inhibition of AMIGO2 reduces bladder cancer cell growth and tumorigenicity in vivo, which was consistent with the in vitro results.
Knockdown of AMIGO2 inhibits tumorigenicity of bladder cancer cells in vivo. A The image of nude mice and tumors. B The volume of tumors, detected for 36 days. The results are presented as the means ± SEM of values (n = 8). Statistical significance was calculated using the Student’s t-tests. (*p < 0.05, **p < 0.01, ***p < 0.001)
Identification of differentially expressed genes (DEGs) and molecular function and pathway enrichment analysis
As the volcano plots illustrated, after data integration, gene expression profiles from RNA Sequencing identified 917 differentially expressed genes. Among all the DEGs, 627 genes were upregulated and 290 were downregulated in shAMIGO2 compared with the shCtrl in T24 (Fig. 4A), grounded on the cut-off criteria (|logFC|> 2, Padj < 0.01). DEGs were selected for integrated analysis. In order to investigate the molecular function and biology pathways of the DEGs, GO and KEGG analysis were performed. GO (Gene Ontology) includes molecular function, biological process and cellular component (Fig. 4B-D). Enriched KEGG pathways of the DEGs are shown in Fig. 4E, including PPAR signaling pathway and some other pathways.
Differentially expressed genes (DEGs) and pathways analyzed by bioinformatics. A Identification of differentially expressed genes. Volcano plot of gene expression profiles. Red/blue symbols classify the upregulated/downregulated genes according to the criteria: |logFC|> 2 and adjusted p-value. B Molecular function, which were significantly enriched in lipid transporter activity, cytokine binding and long-chain fatty acid transporter activity; C Biological process, which were significantly enriched in multicellular organismal homeostasis, extracellular structure organization and synapse organization; D Cellular component, which were significantly enriched in synaptic membrane, extracellular matrix and receptor complex. E KEGG pathways in which DEGs were significantly enriched. There were 8 pathways in the relation graph, including oxytocin signaling pathway, adrenergic signaling in cardiomyocytes, platelet activation, ARVC, HCM, DCM, PPAR signaling pathway and ABC transporters. Size of the dots: Count of DEGs enriched in corresponding GO and KEGG classification
Protein–Protein Interaction (PPI) network construction, hub genes and survival analysis
The PPI network was constructed based on the SRTING database. A total of 174 proteins were obtained from the DEGs, including 136 nodes and 234 edges (Fig. 5A). In the network, nodes with top 10 highest degrees were ZAP70, AKR1C1, MAP2K6, SCN2A, AGMAT, CBLB, AKR1C3, TLR3, SCN3A and AZIN2. These genes were considered as hub genes. The information of the 10 hub genes is shown in Table 4, including full gene names and primary functions. Total 10 hub genes were obtained from PPI network. The Kaplan–Meier survival analysis (Fig. 5B-E) shows that four hub genes associated with survival were ZAP70, AGMAT, AKR1C1 and AKR1C3.
Construction of PPI network and relationship between hub genes and prognosis of bladder cancer. A Modules with relatively high score selected from the protein–protein interaction network. The PPI network contains 136 nodes and 234 edges. ZAP70, AKR1C1, MAP2K6, SCN2A, AGMAT, CBLB, AKR1C3, TLR3, SCN3A and AZIN2 were considered as hub genes. B-E Four genes significantly related to survival. The red line indicates the group with higher expression of this gene, and the blue line indicates the group with lower expression
Overexpression of PPAR-γ reverted the inhibitory effect of AMIGO2 in bladder cancer cells
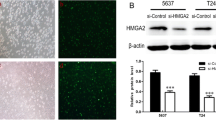
To explore the molecular mechanism of AMIGO2 function in bladder cancer cells, the target of AMIGO2 was predicted using bioinformatics analysis. According to the results of RNA-Seq, PPAR-γ was identified as the target of AMIGO2 (Fig. 4E). PPAR-γ is overexpressed in bladder cancer cell lines T24 and 5637, but when AMIGO2 is knocked down in T24 and 5637, the expression level of PPAR-γ is also decreased (Fig. 6A). Meanwhile, the results of CCK-8 and wound-healing assay both indicated that overexpression of PPAR-γ reverted the inhibitory effect of AMIGO2 on proliferation, migration and cell cycle of bladder cancer cells (Fig. 6B-D). These results suggested that knockdown of AMIGO2 suppresses proliferation and migration by regulating PPAR-γ in bladder cancer.
Overexpression of PPAR-γ reverted the inhibitory effect of AMIGO2 in bladder cancer cells. Experimental grouping: shAMIGO2 + PPAR-γ and Negative Control. A The expression level of PPAR-γ in untreated T24, 5637 and SV-HUC-1 cell lines; the expression level of PPAR-γ in shAMIGO2 + PPAR-γ and Negative Control group of bladder cancer cell lines T24 and 5637. Measured by Western Blot. β-Actin served as the loading control. B Overexpression of PPAR-γ reverted the inhibitory effect of AMIGO2 on proliferation of bladder cancer cells, detected by CCK-8. C Overexpression of PPAR-γ reverted the inhibitory effect of AMIGO2 on migration of bladder cancer cells, assessed by wound healing assay. D Overexpression of PPAR-γ reverted the inhibitory effect of AMIGO2 on cell cycle of bladder cancer cells, analyzed by flow cytometry
Discussion
Previous researches into various genes associated with malignancies has provided valuable insights into the underlying mechanisms of bladder cancer. Studies have indicated that the aberrant expression of specific genes plays a pivotal role in driving the progression of distinct tumor subtypes. In the present study, we found that AMIGO2 was upregulated in bladder cancer cells and tissues, and it could promote the proliferation, migration and tumorigenicity. To explore the mechanism of AMIGO2-induced cell proliferation, we investigated potential targets of AMIGO2 and found that PPAR-γ is targeted by AMIGO2 and is essential for the bio-function of bladder cancer.
The unconstrained cellular proliferation observed in cancer is mainly attributed to the cell cycle deregulation [17]. DNA damage targets two cell cycle checkpoints, namely the G1/S and G2/M. DNA damage triggers a regulatory program that enforces cell cycle at a designated checkpoint, either until the damage is repaired or the cells move towards apoptosis [18]. Arresting cells in G0/G1 phase presents an opportunity for certain cells to either undergo repairing or move towards apoptosis. The flow cytometry assay in our study showed that suppression of AMIGO2 could inhibit its proliferative effects through blockage of cell cycle progression and arrest BCa cells in G1 phase. In many cases, an arrest can lead to senescence or apoptosis [19]. Although few studies have described the relationship between AMIGO2 and cell cycle before, there are some pathways that regulate cell cycle indirectly enriched by our KEGG analysis, but the particular pathways and molecules are still under research.
GO term enrichment analysis showed that the DGEs were significantly enriched in extracellular matrix, lipid transporter activity and cell–cell adhesion via plasma-membrane adhesion molecules, suggesting that some of the DEGs might be involved in cell adhesion and migration. As stated in previous studies, PCDH1 (Protocadherin 1) mediated cell–cell adhesion through homotypic interactions [20], CELSR1 (cadherin EGF LAG seven-pass G-type receptor 1) regulated endothelial adherens junctions and directed cell rearrangements during valve morphogenesis [21], and PCDHGA3 worked as one of the cell adhesion molecules in human ischemic cardiomyopathy [22]. Cell adhesion participants in stimulating signals that regulate cell migration, cell cycle, and cell survival [23]. Cell adhesiveness is generally reduced across different types of human cancers. AMIGO2, as reported before, is also involved in cell adhesion and/or migration [5, 24, 25]. Changes of these molecules might be resulting in the reduction of adhesion between cells and promoting the migration of tumor cells. Correspondingly, the wound healing assay in our study also proved that AMIGO2 could promote the migration of BCa cells.
Subsequently, we explored the interaction of DEGs. A large and complex interactome network was established, suggesting intricate links among those DEGs. Moreover, 10 hub genes were identified in total, 4 of them were related to survival, namely ZAP70, AGMAT, AKR1C1 and AKR1C3. ZAP70 was reported to be involved in the development of chronic lymphocytic leukemia [26]. AGMAT could promote the lung adenocarcinoma tumorigenesis by activating the NO-MAPKs-PI3K/Akt pathway [27]. AKR1C1 was proved to be related with the invasive potential and drug resistance of metastatic bladder cancer cells [28]. AKR1C3, comes from the same family as AKR1C1, has not been proved to have similar functions as AKR1C1 in bladder cancer, but it is often overexpressed in prostate cancer tissues and cell lines [29]. Interestingly, AKR1C3 catalyzes the formation of prostaglandin (PG) F2α and 11β-PGF2α from PGH2 and PGD2, respectively [30]. The PGF2α and 11β-PGF2α can inactivate PPAR-γ and has anti-proliferative effects [31].
PPAR-γ is a member of the nuclear receptor superfamily, it participates in multiple physiological and pathological processes. Extensive researches have revealed the relationship between PPAR-γ and various tumors. However, the expression level and functions of PPAR-γ seem to be controversial [11]. Some studies have indicated that PPAR-γ acts as a tumor promoter [32, 33]. Downregulation of PPAR-γ has been shown to inhibit the growth of cancer cells, suggesting a tumor-promoting effect for PPAR-γ [33, 34]. Nevertheless, some other researches illuminated that PPAR-γ serves as a tumor inhibitor, PPAR-γ suppressed cell growth and invasiveness by blocking cell cycle and stimulating apoptosis and differentiation [7, 35]. In our study, we proved that overexpression of PPAR-γ in AMIGO2-repressed cells could revert the inhibitory effect of shAMIGO2 on bladder cancer cells. Therefore, we consider PPAR-γ as a tumor promoter in bladder cancer.
To the extent of our current understanding, this is the first study determining the very “new” gene AMIGO2, which promotes the proliferation, migration and tumorigenicity in BCa. Mechanistically, we identified the PPAR-γ as a principal molecular subject to modulation by AMIGO2 within bladder cancer cell populations. Interestingly, we found that overexpression of PPAR-γ in AMIGO2-repressed cells could revert the inhibitory effect of shAMIGO2 on bladder cancer cells. As a consequence, it is identified that downregulating AMIGO2 inhibits bladder cancer cells proliferation and tumorigenicity by suppressing PPAR-γ, and that PPAR-γ is essential for AMIGO2-mediated cell proliferation in bladder cancer.
Conclusion
AMIGO2 is overexpressed in bladder cancer cells and tissues. Knockdown of AMIGO2 suppresses bladder cancer cell proliferation and migration. These processes might be regulated by PPAR-γ signaling pathway.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. https://doi.org/10.1038/nature12965.
Boehm BE, Svatek RS. Novel therapeutic approaches for recurrent nonmuscle invasive bladder cancer. Urol Clin North Am. 2015;42(2):159–68. https://doi.org/10.1016/j.ucl.2015.02.001.
Kuja-Panula J, Kiiltomäki M, Yamashiro T, Rouhiainen A, Rauvala H. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J Cell Biol. 2003;160(6):963–73. https://doi.org/10.1083/jcb.200209074.
Rabenau KE, O’Toole JM, Bassi R, Kotanides H, Witte L, Ludwig DL, et al. DEGA/AMIGO-2, a leucine-rich repeat family member, differentially expressed in human gastric adenocarcinoma: effects on ploidy, chromosomal stability, cell adhesion/migration and tumorigenicity. Oncogene. 2004;23(29):5056–67. https://doi.org/10.1038/sj.onc.1207681.
Fontanals-Cirera B, Hasson D, Vardabasso C, Di Micco R, Agrawal P, Chowdhury A, et al. Harnessing BET inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene. Mol Cell. 2017;68(4):731-744 e739. https://doi.org/10.1016/j.molcel.2017.11.004.
Sikka S, Chen L, Sethi G, Kumar AP. Targeting PPARgamma signaling cascade for the prevention and treatment of prostate cancer. PPAR Res. 2012;2012:968040. https://doi.org/10.1155/2012/968040.
Lee JJ, Drakaki A, Iliopoulos D, Iliopoulos D, Struhl K, Struhl K. MiR-27b targets PPARγ to inhibit growth, tumor progression and the inflammatory response in neuroblastoma cells. Oncogene. 2012;31:3818–25. https://doi.org/10.1038/onc.2011.543.
Knutson SK, Chyla BJ, Amann JM, Amann JM, Bhaskara S, Bhaskara S, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J . 2008;27(7):1017–28. https://doi.org/10.1038/emboj.2008.51.
Bhaskara S, Knutson SK Fau, Jiang G, Jiang G, Chandrasekharan MB, Chandrasekharan MB, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18(5):436–47. https://doi.org/10.1016/j.ccr.2010.10.022.
Peng T, Wang G, Cheng S, Xiong Y, Cao R, Qian K, et al. The role and function of PPARgamma in bladder cancer. J Cancer. 2020;11(13):3965–75. https://doi.org/10.7150/jca.42663.
Zlobec I, Suter G, Perren A, Lugli A. A next-generation tissue microarray (ngTMA) protocol for biomarker studies. J Vis Exp. 2014;91:51893. https://doi.org/10.3791/51893.
Wang B, Xu D, Yu X, Ding T, Rao H, Zhan Y, et al. Association of intra-tumoral infiltrating macrophages and regulatory T cells is an independent prognostic factor in gastric cancer after radical resection. Ann Surg Oncol. 2011;18(9):2585–93. https://doi.org/10.1245/s10434-011-1609-3.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–8. https://doi.org/10.1038/nprot.2008.73.
Cumashi A, Tinari N, Rossi C, Lattanzio R, Natoli C, Piantelli M, et al. Sunitinib malate (SU-11248) alone or in combination with low-dose docetaxel inhibits the growth of DU-145 prostate cancer xenografts. Cancer Lett. 2008;270(2):229–33. https://doi.org/10.1016/j.canlet.2008.05.007.
Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. https://doi.org/10.1186/s13059-016-0881-8.
Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–66. https://doi.org/10.1038/nrc2602.
Sitko JC, Yeh B, Kim M, Zhou H, Takaesu G, Yoshimura A, et al. SOCS3 regulates p21 expression and cell cycle arrest in response to DNA damage. Cell Signal. 2008;20(12):2221–30. https://doi.org/10.1016/j.cellsig.2008.08.011.
Lagunas-Cruz MDC, Valle-Mendiola A, Trejo-Huerta J, Rocha-Zavaleta L, Mora-Garcia ML, Gutierrez-Hoya A, et al. IL-2 induces transient arrest in the G1 phase to protect cervical cancer cells from entering apoptosis. J Oncol. 2019;2019:7475295. https://doi.org/10.1155/2019/7475295.
Faura Tellez G, Willemse BW, Brouwer U, Nijboer-Brinksma S, Vandepoele K, Noordhoek JA, et al. Protocadherin-1 localization and cell-adhesion function in airway epithelial cells in asthma. PLoS One. 2016;11(10):e0163967. https://doi.org/10.1371/journal.pone.0163967.
Tatin F, Taddei A, Weston A, Fuchs E, Devenport D, Tissir F, et al. Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell. 2013;26(1):31–44. https://doi.org/10.1016/j.devcel.2013.05.015.
Ortega A, Gil-Cayuela C, Tarazon E, Garcia-Manzanares M, Montero JA, Cinca J, et al. New Cell Adhesion Molecules in Human Ischemic Cardiomyopathy. PCDHGA3 Implications in decreased stroke volume and ventricular dysfunction. PLoS One. 2016;11(7):e0160168. https://doi.org/10.1371/journal.pone.0160168.
Huang S, Ingber DE. The structural and mechanical complexity of cell-growth control. Nat Cell Biol. 1999;1(5):E131–8. https://doi.org/10.1038/13043.
Kanda Y, Osaki M, Onuma K, Sonoda A, Kobayashi M, Hamada J, et al. Amigo2-upregulation in tumour cells facilitates their attachment to liver endothelial cells resulting in liver metastases. Sci Rep. 2017;7:43567. https://doi.org/10.1038/srep43567.
Chen J, Gu L, Ni J, Hu P, Hu K, Shi YL. MiR-183 regulates ITGB1P expression and promotes invasion of endometrial stromal cells. Biomed Res Int. 2015;2015:340218. https://doi.org/10.1155/2015/340218.
Carabia J, Carpio C, Abrisqueta P, Jimenez I, Purroy N, Calpe E, et al. Microenvironment regulates the expression of miR-21 and tumor suppressor genes PTEN, PIAS3 and PDCD4 through ZAP-70 in chronic lymphocytic leukemia. Sci Rep. 2017;7(1):12262. https://doi.org/10.1038/s41598-017-12135-7.
Zhu HE, Yin JY, Chen DX, He S, Chen H. Agmatinase promotes the lung adenocarcinoma tumorigenesis by activating the NO-MAPKs-PI3K/Akt pathway. Cell Death Dis. 2019;10(11):854. https://doi.org/10.1038/s41419-019-2082-3.
Matsumoto R, Tsuda M, Yoshida K, Tanino M, Kimura T, Nishihara H, et al. Aldo-keto reductase 1C1 induced by interleukin-1beta mediates the invasive potential and drug resistance of metastatic bladder cancer cells. Sci Rep. 2016;6:34625. https://doi.org/10.1038/srep34625.
Doig CL, Battaglia S, Khanim FL, Bunce CM, Campbell MJ. Knockdown of AKR1C3 exposes a potential epigenetic susceptibility in prostate cancer cells. J Steroid Biochem Mol Biol. 2016;155(Pt A):47–55. https://doi.org/10.1016/j.jsbmb.2015.09.037.
Liu C, Yang JC, Armstrong CM, Lou W, Liu L, Qiu X, et al. AKR1C3 promotes AR-V7 protein stabilization and confers resistance to AR-targeted therapies in advanced prostate cancer. Mol Cancer Ther. 2019;18(10):1875–86. https://doi.org/10.1158/1535-7163.MCT-18-1322.
Julian CD, et al. The aldo-keto reductase AKR1C3 is a novel suppressor of cell differentiation that provides a plausible target for the non-cyclooxygenase-dependent antineoplastic actions of nonsteroidal anti-inflammatory drugs. Cancer Res. 2003;63:505–12.
Lee JJ, Drakaki A, Iliopoulos D, Struhl K. MiR-27b targets PPARgamma to inhibit growth, tumor progression and the inflammatory response in neuroblastoma cells. Oncogene. 2012;31(33):3818–25. https://doi.org/10.1038/onc.2011.543.
Hsu HT, Sung MT, Lee CC, Kuo YJ, Chi CW, Lee HC, et al. Peroxisome proliferator-activated receptor gamma expression is inversely associated with macroscopic vascular invasion in human hepatocellular carcinoma. Int J Mol Sci. 2016;17(8):1226. https://doi.org/10.3390/ijms17081226.
Zaytseva YY, Wang X, Southard RC, Wallis NK, Kilgore MW. Down-regulation of PPARgamma1 suppresses cell growth and induces apoptosis in MCF-7 breast cancer cells. Mol Cancer. 2008;7:90. https://doi.org/10.1186/1476-4598-7-90.
Ota K, Ito K, Suzuki T, Saito S, Tamura M, Hayashi S, et al. Peroxisome proliferator-activated receptor gamma and growth inhibition by its ligands in uterine endometrial carcinoma. Clin Cancer Res. 2006;12(14 Pt 1):4200–8. https://doi.org/10.1158/1078-0432.CCR-05-1833.
Han D, Xiong B, Zhang X, Chen C, Yao Z, Wu H, et al. Effects and mechanisms of AMIGO2 in proliferation migration and drug resistance of bladder cancer. 2020. https://doi.org/10.21203/rs.3.rs-34249/v1.
Acknowledgements
A preprint has previously been published [36].
Funding
This work was supported by the Science and Technology project of Chengguan District, Lanzhou city, Gansu province Science and Technology Bureau (Project number: 2017KJGG0052), the Cuiying Graduate Supervisor Applicant Training Program of Lanzhou University Second Hospital (201704), the Gansu Health Industry Research Project (GSWSKY2017-10), the Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital (Project number: CY2017-BJ16, Doctoral supervisor training program), and the National Nature Science Foundation of China (NO: 81372732&30800206). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.-q.T. conceived and designed the experiments. D.-l.H., B.X., X.-x.Z., C.-h.C., Z.-q.Y. and H.W. performed the experiments and analyzed the data. B.X., J.-l.C., J.-p.L. and P.L. did bioinformatics analysis and provided statistical support. D.-l.H. prepared the manuscript. Z.-p.W. helped write the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This project was approved by the Institutional Research Ethics Committee of the Second Hospital of Lanzhou University (No. 2019A-119 and No. D2019-171). The ethical approval and the informed consent were supplied in attachment files.
Consent for publication
Included.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Han, D., Xiong, B., Zhang, X. et al. Knockdown of AMIGO2 suppresses proliferation and migration through regulating PPAR-γ in bladder cancer. Hereditas 161, 21 (2024). https://doi.org/10.1186/s41065-024-00325-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41065-024-00325-z