Abstract
Background
The Drosophila hindgut is commonly used model for studying various aspects of organogenesis like primordium establishment, further specification, patterning, and morphogenesis. During embryonic development of Drosophila, many transcriptional activators are involved in the formation of the hindgut. The transcription factor Orthopedia (Otp), a member of the 57B homeobox gene cluster, is expressed in the hindgut and nervous system of developing Drosophila embryos, but due to the lack of mutants no functional analysis has been conducted yet.
Results
We show that two different otp transcripts, a hindgut-specific and a nervous system-specific form, are present in the Drosophila embryo. Using an Otp antibody, a detailed expression analysis during hindgut development was carried out. Otp was not only expressed in the embryonic hindgut, but also in the larval and adult hindgut. To analyse the function of otp, we generated the mutant otp allele otpGT by ends-out gene targeting. In addition, we isolated two EMS-induced otp alleles in a genetic screen for mutants of the 57B region. All three otp alleles showed embryonic lethality with a severe hindgut phenotype. Anal pads were reduced and the large intestine was completely missing. This phenotype is due to apoptosis in the hindgut primordium and the developing hindgut.
Conclusion
Our data suggest that Otp is another important factor for hindgut development of Drosophila. As a downstream factor of byn Otp is most likely present only in differentiated hindgut cells during all stages of development rather than in stem cells.
Similar content being viewed by others
Background
The Drosophila embryonic hindgut is a single-layered ectodermally derived epithelium surrounded by visceral musculature [1,2,3]. It arises from a group of cells at the posterior part of the blastoderm stage embryo referred to as the hindgut primordium [1]. The hindgut primordium is a ring of about 200 blastoderm cells that is internalised during gastrulation to form a short, wide sac. In a relatively short time this epithelium sac is transformed into a long tube containing approximately 700 cells [1, 4, 5]. The growth of the hindgut starting at stage 12 is not due to cell divisions, but a twofold endoreplication [6] that leads to an increase in cell size, and as a consequence total length of the hindgut [7, 8]. During this process, the developing hindgut becomes subdivided along the anterior posterior (AP) axis and the dorsoventral (DV) axis. Along the AP axis, the hindgut forms three morphologically distinct regions: the small intestine, large intestine, and rectum [5, 8, 9]. The small intestine is the most anterior part of the hindgut and is connected to the posterior midgut, whereas the large intestine is the central part of the hindgut and forms three distinct regions along the DV axis. The dorsal and ventral regions constitute the outer and inner portions of the hindgut loop, respectively. Two rows of boundary cells are organised between these two regions and as two rings at the anterior and posterior borders of the large intestine [8, 10, 11]. The most posterior-most portion of the hindgut is the rectum, which connects to the anal pads.
Several genes are required to establish the hindgut primordium and to pattern the hindgut along the AP axis. At the blastoderm stage a group of posterior cells, called the proctodeal primordium (Campos-Ortega and Hartenstein, 1997) [1], will later on give rise to the hindgut. In these cells the transcription factor Tailless (Tll) [12] is expressed and subsequently activates other transcription factors like Brachyenteron (Byn) [13], Fork head (Fkh) [14] and Bowel (Bowl) [15] as well as the signalling protein Wingless (Wg) [16], which are all necessary for hindgut development. The transcription factor Caudal (Cad) [17] is also expressed in the proctodeal ring, but independently of Tailless. Tll [18] and Wg [9, 19, 20] are necessary to establish the primordium, whereas Cad is necessary for the internalisation of the hindgut primordium later on [21]. Proper gene expression in and maintenance of the large intestine requires byn, Dichaete (D), raw, lines (lin) and mummy (mmy), while bowl and drumstick (drm) are required for gene expression in and maintenance of the small intestine [5, 19, 22,23,24,25,26].
The Drosophila T-box gene brachyenteron (byn) is expressed in the ring of cells that internalise to form the embryonic hindgut and expression is maintained in the hindgut throughout embryogenesis [13]. In byn mutants the hindgut is shortened due to apoptosis and the large intestine is missing [11, 22]. The Drosophila homeobox gene orthopedia (otp) is also expressed in the hindgut, anal pads and along the central nervous system [27]. It is located in 57B region of the second chromosome in close vicinity to the other homeobox genes Drosophila retinal homeobox (Drx) [28, 29] and homeobrain (hbn) [30]. In the hindgut, otp is directly activated by byn in a dose-dependent manner via multiple binding sites present in a regulatory element of otp [31].
Otp is highly conserved through evolution and has been identified in most multicellular organisms. Among these are several invertebrates such as sea urchins [27], the mollusc Patella vulgata [32], the annelid Platynereis dumerilii [33] and several vertebrates such as zebrafish, that have two genes namely otp1 and otp2 [34], chicken [27, 35], mouse [27] and human [36]. Otp genes of vertebrates have a major function in the development of the hypothalamic neuroendocrine system (see [37] for review).
The function of otp during Drosophila development has been unknown so far as no mutants have been described. In the present study, we show that otp is required for proper hindgut development in Drosophila. We generated one otp allele by ends-out gene targeting and isolated two additional otp alleles in an EMS-mutagenesis screen. All three otp alleles are characterised by a dramatically reduced hindgut lacking the complete large intestine. This reduction in hindgut length is due to apoptosis in the hindgut primordium and the developing hindgut.
Results
Hindgut and nervous system specific transcripts of the otp gene
The expression of the otp gene during embryonic development was first described by Simeone et al. (1994) [27]. The gene is expressed first in the hindgut primordium, then in the hindgut and anal pads as well as in the ventral nerve cord and in the brain of the Drosophila embryo. With the help of cDNAs and ESTs which have been analysed over the years nine different transcript forms of otp are known today (Flybase FB2020_03, [38]). They basically fall into two different classes, some which have exons 1 and 2 spliced to exon 4, here otp-RE is an example, and others which have exon 3 spliced to exon 4, here otp-RC is one representative (Flybase FB2020_03, [38]) (Fig. 1a). A 2.3 kb transcript (referred to as otp-RC by Flybase) can first be detected in 3–6 h old embryos. In 9–12 h old embryos an additional 2.9 kb transcript (referred to as otp-RE by Flybase) can be detected. The expression of both transcripts remains throughout all embryonic and larval stages.
Hindgut and nervous system specific transcripts of the orthopedia (otp) gene. a Genomic organisation of the otp gene with exons indicated as gray boxes and the regulatory region for the otp hindgut expression with the Byn binding sites (hindgut module) [31]. Below the two different transcripts (otp-RE and otp-RC) are shown (gray, untranslated region; blue, translated region; yellow, homeobox). The specific genomic probes (otp 1 and otp 2) used to detect otp-RE and otp-RC by in situ hybridisation are indicated. b-j Embryonic expression pattern of the different otp transcripts. The stages shown (9, 10, and 14) were determined according to Campos-Ortega and Hartenstein (1997) [1], all views are lateral views, anterior is to the left. b-d Whole-mount in situ hybridisation performed with an otp cDNA probe against exons 3–9 detecting all transcript forms (otp-RC). The boundary between stronger anterior expression and weaker posterior expression in the nervous system is indicated by a blue arrowhead, the weaker expression in the rectum compared with the rest of the hindgut by a black arrowhead in d. e-g In situ hybridisation performed with a probe against exon 1 and 2 of the otp gene (otp 1). h-j In situ hybridisation performed with a probe against exon 3 of the otp gene (otp 2)
Because the two otp transcripts show different temporal expression we contemplated whether they also differ in their spatial expression. Since the two otp transcripts vary in the 5′ part, exons 1 and 2 are specific to the larger 2.9 kb transcript, exon 3 is specific to the smaller 2.3 kb transcript, and each transcript can be detected with a specific probe.
After analysing whole mount in situ hybridisations of embryos with a probe detecting exons 3–9 sequences (probe otp-RC) that are present in both transcripts, the otp expression was mostly consistent with that described by Simeone et al. (1994) [27]. The earliest expression was visible in the hindgut primordium, the proctodeum, at stage 9 (Fig. 1b), an additional otp expression in the nervous system begins at stage 10 (Fig. 1c) and otp is expressed in the hindgut, anal pads, ventral nerve cord and brain at stage 14 (Fig. 1d). Whereas Simeone et al. (1994) [27] detected no expression of otp in the rectum, we detected otp expression in this small portion at the most posterior part of the hindgut, albeit at a lower level than in the other parts of the hindgut (Fig. 1d, black arrowhead). In the nervous system, there was a clear border with a higher level of otp expression in the five most anterior segments of the ventral nerve cord and a lower level of otp expression in the posterior segments of the ventral nerve cord (Fig. 1d, blue arrowhead indicating the border).
Using a probe against exons 1 and 2 of the otp gene (probe otp 1), no expression was detected in stage 9 embryos (Fig. 1e). At stage 10, otp expression was only visible in the nervous system, but not in the hindgut primordium (Fig. 1f). At stage 14, otp expression was again only present in the ventral nerve cord and brain (Fig. 1g).
With a probe against exon 3 of the otp gene (probe otp 2), the complementary expression pattern can be detected. At stage 9 otp expression occurs in the hindgut primordium (Fig. 1h), and at stage 10, otp expression continues embryos also in the hindgut primordium, but not in the nervous system (Fig. 1i). At stage 14, otp expression can be detected in the hindgut and anal pads, but not in the ventral nerve cord and brain (Fig. 1j).
These results indicate that the smaller 2.3 kb otp transcript is specifically expressed in the hindgut primordium, embryonic hindgut and anal pads, whereas the expression of the larger 2.9 kb otp transcript is restricted to the embryonic nervous system, where it is expressed in the ventral nerve cord and brain.
Otp expression during embryonic development
In order to analyse the Otp protein expression at a higher resolution, we produced an antibody against the Otp-C-terminus in guinea pigs. As seen in the case of the otp mRNA, protein expression started in the proctodeum at stage 10, which was detectable in nuclei as expected (Fig. 2a). During germband retraction at stage 12 the developing hindgut already had the shape of a small tube and the anal pads were visible (Fig. 2b). When the germband retraction was finished at stage 14 expression was seen in the hindgut and the anal pads, but in addition also in the brain and ventral nerve cord (Fig. 2c). One major difference between the protein expression and mRNA expression was detectable in the ventral nerve cord, where the protein is only expressed up to segment A2 (Fig. 2c, white arrowhead) and not in the more posteriorly located segments. This was even better visible in a ventral view of the nerve cord at stage 15 (Fig. 2d, white arrowhead). In the embryonic brain, expression was seen in three discrete expression domains per hemisphere, labeled P1, P2, and D1 from anterior to posterior with respect to the neuraxis (Fig. 2e). Compared to the supraesoghageal commissure seen by the Nrt staining (white arrowhead), P1 and P2 were located in the protocerebrum and D1 in the deutocerebrum. The expression domains consisted of 10 (P1), 3 (P2) and 5 (D1) neurons (Fig. 2e).
Expression of Otp during embryonic development. Laser confocal images of Drosophila embryos labeled with an anti-Otp antibody (green). Stages were determined according to Campos-Ortega and Hartenstein (1997) [1] as indicated in the figure. a-c Lateral views of embryos, the anterior ends are to the left. In c the expression border in the ventral nerve cord is shown by an arrowhead. d Ventral view of the embryonic nerve cord, segments T1-A2 are labeled and the border of the nervous system expression is again indicated by an arrowhead. e Dorsal view of an embryonic brain stained with anti-Otp (green) and anti-Nrt antibodies (red) to visualize the primary neurons, the Otp expression domains are labelled P1 (protocerebrum), P2 (protocerebrum) and D1 (deutocerebrum). The supraesoghageal commissure is indicated by an arrowhead
Expression of Otp in the embryonic, larval and adult hindgut
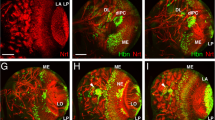
Like otp mRNA, Otp protein was expressed in the embryonic hindgut and anal pads at embryonic stage 14 (Fig. 3a). Byn is a direct regulator of otp and sufficient to induce otp expression [31]. The enhancer trap line bynapro which mimics Byn protein expression [10] was therefore used to investigate in which parts of the hindgut Otp and Byn are co-localised. Otp and Byn showed co-localisation in large parts of the hindgut primordium (data not shown) and along the complete large intestine (Fig. 3a-c), whereas in the small intestine, only Byn was expressed. Both Otp and Byn are also co-expressed in the rectum, but as for the otp mRNA the level of Otp protein in this part of the hindgut was lower than in the large intestine and anal pads (Fig. 3a-c). By contrast, the level of Byn expression did not differ between the different parts of the hindgut. Byn and Otp were also co-localised in the anal pads with an additional outer ring of cells only expressing Byn (Fig. 3a-c). Both the larval and the adult hindgut were subdivided along the anterior-posterior axis into three segments: the pylorus, ileum, and rectum. The most anterior part, the pylorus, which is also called the hindgut proliferation zone (HPZ), is an important region for the formation of the adult hindgut, because the cells of the larval hindgut undergo apoptosis during metamorphosis and are replaced by cells of the HPZ to form the adult hindgut [39]. In our study, in the larval hindgut, Otp and Byn were co-expressed in most parts (Fig. 3d-f). The only exception here was again the most anterior region, the pylorus, where only Byn was expressed (Fig. 3e), but not Otp (Fig. 3d). The co-expression of Otp and Byn was also visible in the adult hindgut (Fig. 3g-i). Here, as in the larval hindgut, Otp was only expressed in the ileum and rectum, whereas Byn was additionally expressed in the pylorus. The expression of both factors suggested a role not only in embryonic and larval hindgut development, but also in maintenance of the adult hindgut as well.
Embryonic, larval and adult hindgut expression pattern of the Otp protein. Anti-Otp antibody staining in green (a, d, g), anti-β-Galactosidase antibody staining of bynapro in red (b, e, h) and merging of both stainings (c, f, i). In all views, anterior is to the left, embryos are shown as lateral views. a-c stage 16 embryo, d-f larval hindgut, g-i adult hindgut. The borders between different structures are indicated by white arrowheads. AP, anal pads; HG, hindgut; IL, ileus; LI, large intestine; PY, pylorus; RE, rectum; SI, small intestine
Generation of otp alleles
So far, no otp alleles have been described in previous studies. Thus, in order to analyse the function of otp in Drosophila development, we generated otp alleles by two different approaches. In ends-out gene targeting [40,41,42], we generated the mutant otp allele otpGT (Fig. 4a). In this mutant, the translation start site resulting from the translation of the hindgut specific transcript and most of the homeobox of the otp gene were missing thereby completely inactivating the otp function. The correct gene targeting event was verified by PCR amplification of both ends of the targeting construct.
Generation of the otp GT allele and molecular analysis of the otp1024 allele. a At the top, the genomic organization of the otp gene is shown with exons indicated as gray boxes. The otp gene targeting construct in the targeting vector pW25 (otpGT-pW25) is shown below (FRT, FLP recombination target sequences; I-site, I-SceI recognition site; Whs, white gene with hsp70 promoter). Further down the otp genomic region after the gene targeting event is indicated (otpGT); here part of exon 4 (containing the translation start) and the complete exon 5 (containing most part of the homeobox) are replaced by the white gene of the targeting construct. b Nucleotide and amino acid sequences in wild-type (here otp-RC as an example) and mutant DNA. EMS induced a G to A transition in the coding region of otp1024 leading to the formation of a stop codon. c Schematic overview of the wild-type Otp protein with the localisation of the homeodomain (yellow) in comparison to the truncated mutant protein of the otp1024 allele
In a previous EMS-mutagenesis screen [43] for mutants of the 57B region, 33 lethal mutations representing 16 complementation groups were isolated. By complementation analysis using the gene targeting strain otpGT, we identified two mutant strains (1024 and 13064) not complementing the otpGT allele and also each other, suggesting that they represent otp alleles. To analyse these alleles at the molecular level, all otp exons including the exon-intron boundaries of both strains were PCR amplified and sequenced. Unfortunately, no sequence alterations were identified in strain 13064, but in strain 1024, a single G to A transition in exon 6 induced a change from a tryptophan codon into a stop codon at amino acid position 67 in the carboxy terminus of the homeodomain (Fig. 4b, c). These results verified that the mutant strains 1024 and 13064 represent otp alleles, from now on called otp1024 and otp13064.
Lethality and phenotype of otp alleles
To determine the strength of each otp allele, we analysed their lethality periods by comparing the lethality rates of homozygous otp1024, otp13064 and otpGT mutant animals with those of wild-type animals. All three otp alleles showed embryonic lethality. Furthermore, we investigated the embryonic phenotype that caused that lethality by focusing on the hindgut, the major expression domain of otp. We used Crumbs (Crb) which is expressed on the apical surface of ectodermally derived epithelia [44] as a marker in combination with Otp to detect changes in hindgut length. In the hindgut, Crb labels the anterior and posterior boundary cells of the large intestine as well as the borders between the dorsal and ventral domains of the large intestine [8]. In wild-type embryos, the described expression of Crb in the large intestine was clearly visible, and the expression of Otp in the rectum and anal pads in addition to the large intestine was also observed (Fig. 5a). In contrast to wild-type embryos, anti-Otp-antibody staining revealed that homozygous mutant embryos of all three otp alleles are characterized by a complete loss of Otp expression (Fig. 5b-d). Crb staining shows changes in hindgut length and morphology in homozygous embryos of all three otp alleles (Fig. 5b-d, arrowheads). Compared to the hindgut length of wild-type stage 16 embryos, the hindgut length of homozygous otp embryos is only about one third of wild-type length.
Shortening of the hindgut in otp mutants. Anti-Otp- (red) and anti-Crumbs- (Crb) (green) antibody staining of stage 16 wild-type embryos (a), homozygous otp1024 (b), otp13064 (c) and otpGT (d) embryos. In wild-type embryos (a) the borders of the large intestine (LI) are marked by red arrowheads, and the shortened hindgut of homozygous otp1024, otp13064, and otpGT embryos (b-d) is indicated by white arrowheads. All views are lateral views, anterior is to the left
To analyse the effect of a mutation in otp more specifically, we used an in situ hybridisation to compare the distribution of the otp mRNA from wild-type embryos with otp1024 mutant embryos. Assuming that the point mutation in otp1024 embryos has no effect on the mRNA expression, a loss of expression in certain regions might give a first hint for affected areas in otp mutants. In stage 14 wild-type embryos otp was expressed in the large intestine, rectum and anal pads (Fig. 6a). In contrast to Otp protein otp mRNA could be detected in the remaining part of the hindgut of homozygous otp1024 embryos (Fig. 6d). Compared to wild-type embryos, the anal pads of otp1024 embryos were reduced and not located at the posterior end of the embryo. While the rectum was still present, but the large intestine was missing (Fig. 6d, black arrowhead). Additional markers were used to find out which of the different hindgut regions are affected in otp mutants. Hedgehog (hh) encodes a cell signalling molecule that is required for the maintenance of the small intestine and rectum where it is expressed [8, 9] (Fig. 6b). These two expression domains were not affected in the hindgut of otp1024 embryos but they are moved further together (Fig. 6e, black arrowhead) indicating that the part between these two domains is affected. The AT-rich interaction domain (AID) box gene retained (retn) / dead ringer (dri) was expressed in the boundary cells of the large intestine (Fig. 6c, black arrowheads) and at the border between the large intestine/small intestine and small intestine/midgut [45, 46] (Fig. 6c). No expression of retn / dri could be detected in the boundary cells between the ventral and dorsal region of the large intestine and at the posterior border of the large intestine in homozygous otp1024 embryos (Fig. 6f, black arrowheads) indicating that these cells are missing in otp mutants. The other two otp alleles also showed a similar hindgut phenotype (data not shown) and therefore can be considered as equally strong. Taken together these results revealed that the small intestine and rectum develop quite normally and showed a normal expression pattern of the markers used in homozygous otp mutant embryos, whereas the large intestine was dramatically affected. No expression of large intestine specific genes could be detected, suggesting that this central domain is completely missing resulting in a reduction in hindgut length due to the loss of the large intestine. In addition to the effect in the hindgut, the anal pads were reduced in size and did not reach the posterior end of the embryo.
The otp hindgut phenotype. Whole mount in situ hybridisation of otp (a, d), hh (b, e) and dri (c, f) mRNA in stage 14 wild-type (a-c) and otp1024 mutant embryos (D-F); the probes are indicated on the right, the genotypes of the embryos on the left. In all views, anterior is to the left, all views are lateral views. Compared to wild-type embryos (a), the large intestine is almost completely missing in otp1024 embryos (black arrowhead), and only expression in the rectum and anal pads is visible using an otp probe (d). The expression of hh in the small intestine and rectum seen in wild-type embryos (b) is closer together in otp1024 mutant embryos (black arrowhead) due to the missing large intestine (e). From the expression of dri in the border cells and the boundaries between large intestine/small intestine and small intestine/midgut (c) only the small intestine/midgut expression is left, the other expression domains are missing (f; black arrowheads). AP, anal pads; BC, boundary cells; LI, large intestine; MG, midgut; RE, rectum; SI, small intestine; dri, dead ringer; hh, hedgehog; otp, orthopedia
Apoptosis in the hindgut primordium of otp embryos
The primary cause of the reduced hindgut and anal pads in byn mutants is ectopic cell death in the hindgut primordium [22]. In order to investigate whether the reduced hindgut of otp mutants might also be caused by apoptosis, we analysed the expression of the pro-apoptotic gene reaper (rpr) in homozygous mutant otp embryos (Fig. 7). Rpr RNA is expressed in cells about to undergo apoptosis [47]. During hindgut development of wild-type stage 10 embryos, there were some rpr expressing cells in the hindgut primordium (Fig. 7a, white arrowhead), but apoptosis does not play a role in determining the size or morphology of the embryonic hindgut in wild-type embryos [5]. At the same stage an increasing number of rpr expressing cells were detected in the hindgut primordium of homozygous mutant embryos from all otp mutant alleles, here an otp1024 embryo is shown as an example (Fig. 7A’, white arrowhead). In addition to rpr as a cell death marker, we also used an antibody against the Drosophila effector caspase Dcp-1 [48] and stained all otp mutant strains. Here again an increased amount of Dcp-1 is present in the hindgut primordium of all otp alleles compared to the wildtype (Fig. 7b, white arrowhead), and an otp13064 embryo is shown as an example (Fig. 7B’). These results indicate that the reduced hindgut of otp embryos is caused by elevated apoptosis in the hindgut primordium and developing hindgut.
Apoptosis in wild-type and otp mutant embryos. Whole mount in situ hybridisation of reaper (rpr) mRNA in wild-type (a) and homozygous otp1024 mutant embryos (a’) at stage 10 (white arrowheads). Antibody staining with anti-Otp antibodies (red) and anti-Cleaved Drosophila Dcp-1 antibodies (green) to highlight apoptotic cells in wild-type (b) and otp13064 mutant embryos (b’) at stage 10 (white arrowheads). Stages were determined according to Campos-Ortega and Hartenstein (1997) [1]. All views are lateral views, anterior is to the left
Discussion
In this paper, we analysed the function of the transcription factor Orthopedia during hindgut development. In the embryo, Otp is expressed in the hindgut primordium, the developing hindgut, and the anal pads. This expression is dependent on several upstream regulators, such as lines [5] and byn [31]. Byn directly activates otp through modular binding sites upstream of hindgut specific promoter of otp in a cooperative fashion [31]. Otp is then expressed in the large intestine, rectum and anal pads, unlike Byn, which is also expressed in the small intestine. Byn alone is therefore not sufficient to activate otp; lines expression might also be needed. In the small intestine where byn is expressed, lines is repressed by drumstick preventing otp activation [49]. If lines is overexpressed in the small intestine, the repressive effect of drumstick can be overruled and otp expression can take place (data not shown), which supports the proposed model that lines and byn are necessary for otp expression in the hindgut. Once otp is activated in the embryonic hindgut, its expression continues till the adult stage. The only region where otp in contrast to byn is not expressed is the larval hindgut proliferation zone in the anteriorly located pylorus which supposedly generates hindgut stem cells capable of replacing the larval hindgut cells undergoing apoptosis and building the adult hindgut. The presence of adult hindgut stem cells was questioned by Fox and Spradling (2009) [50], who showed that proliferation only takes place in response to tissue damage. The current view is that all parts of the Drosophila intestinal tract maintain stem cells that could migrate across organ boundaries [51]. Since otp expression was never shown in areas where intestinal stem cells are present, otp is rather expressed in and a marker for differentiated hindgut cells.
To analyse the function of otp, we generated a mutant allele by gene targeting via homologous recombination and using this targeting strain, we identified two additional mutant alleles by complementation among our EMS-induced collection of mutants from the 57B region. In the gene targeting mutant, part of exon 4 and exon 5 were missing resulting in an N-terminal deletion of the otp-PC protein form including most of the homeodomain. In the otp1024 mutant, the N-terminus and most of the homeodomain were present, but the C-terminal part of the protein was missing. In both cases no Otp protein expression was detected since our anti-Otp antibody was directed against the C-terminal part of Otp. In the case of otp13064, we could also not detect a protein with our anti-Otp antibody nor detect a sequence alteration in the coding region. The splice sites were intact, but we cannot rule out the possibility that a cryptic splice site might be generated. The generation of cryptic splice sites by mutations is often the case in human genetic disorders like Neurofibromatosis type I (NFI) [52] or Cystic fibrosis (CF) [53] where such mutations generate pseudo-exons (see [54] for review). Another possibility might be a mutation in a regulatory region of the gene. All otp alleles showed embryonic lethality with a strong hindgut phenotype. The loss of the large intestine led to a dramatic reduction in hindgut length to about one third of the wildtype length. This phenotype is comparable to the byn phenotype, since byn is directly regulating otp [31]. The three transcriptional regulators drm, bowl and lin are required for patterning and cell rearrangements during elongation in the hindgut, but when compared to otp, showed only a reduction to 40% (drm and bowl) or 50% (lin) in the mutant phenotype [5], suggesting that the loss of otp function is much more severe compared to these genes. A gut specific function of otp like seen in Drosophila is not known for otp genes in higher organisms, where an expression in the nervous system and a function in various aspects of nervous system development is known. In Drosophila, otp is also expressed in the ventral nerve cord and the brain. Expression in the nerve cord seems to be post-transcriptionally regulated, since, in contrast to the mRNA expression posterior to segment A2, the Otp protein is not expressed there. This might be due to the regulation via a miRNA as it was seen for other developmental processes (see [55] for review).
The nervous system function of otp in higher organisms has been mainly analysed in various model organisms like zebrafish and mice. Here it was shown that Otp is expressed in the hypothalamus that exerts influence on physiological function in various processes like blood pressure, circadian rhythmicity, energy balance and homeostasis [37]. In zebrafish Otp in the hypothalamus is required in the preoptic area for the production of the neurohypophysial peptide arginine vasotocin [56], for dopaminergic and neuroendocrine cell specification [57, 58], regulation of stress response [59, 60] and through neuropeptide switching that impacts social behavior [61]. In mice, a loss of Otp leads to a progressive impairment of neuroendocrine cells in the hypothalamus, and homozygous mutant animals die soon after birth with a failure of terminal differentiation of neurosecretory cells [62, 63]. Recently, it was shown that a mutation in Otp is associated with obesity and anxiety in mice [64]. The OTP gene from humans was cloned some time ago [36], but only during the last few years, it could be shown that OTP has a high diagnostic value concerning pulmonary neuroendocrine tumours. Even if these tumours accounted for only 1–2% of all lung tumours, their occurrence increased over the last decades [65]. People with poor survival rates showed a strong downregulation of OTP [66]. OTP that is normally located in the nucleus (nOTP) could also be detected in the cytoplasm (cOTP) or not be present at all. Patients with nOTP have a favourable disease outcome, those with cOTP have an intermediate outcome and those with no OTP expression have the worst disease outcome [67] demonstrating the diagnostic value of OTP. Due to these very interesting aspects of Otp function in the nervous system of higher organisms, it would be interesting to analyse the function of Otp during embryonic brain development of Drosophila, as well as later functions of Otp during larval development and in the adult, using our newly generated otp alleles in the future.
Conclusions
Using gene expression analysis and newly generated mutant otp alleles, we showed that the Drosophila homeodomain transcription factor Orthopedia is an important factor for hindgut development. Our findings demonstrate a requirement of otp to build the large intestine of the hindgut and also in the correct formation of the anal pads. This phenotype is caused by apoptosis at the beginning of hindgut development. Otp as a downstream factor of byn is most likely present only in differentiated hindgut cells during all stages of development rather than in stem cells.
Methods
Whole-mount in situ hybridisation
To synthesise DIG-labeled DNA probes the following DNAs were used: A dri/rtn cDNA clone (LD35748, BDGP), a hh cDNA clone [68], an otp cDNA clone containing exons 3–9 (otp-RC), a 1.6 kb genomic otp clone containing exons 1 and 2, an otp fragment containing exon 3 amplified with the primers 10,036–1 (5′-TCGCACCTCGGTTTCTTCTAC-3′) and 10,036–2 (5′-GGCAATAGTTATTCCACCGATC-3′), a 0,8 kb fragment of the reaper gene amplified with the primers rpr1 (5′-GTCATTGAATAAGAGAGACACC-3′) and rpr2 (5′-GCAATTTTTAGCCAACTTCGAC-3′) and a 1.0 kb fragment of the byn gene amplified with the primers byn1 (5′-GTTCCTGGTGGCAAGGCAGAG-3′) and byn2 (5′-AGATGTTGCGGCCACTGCACAC-3′). In situ hybridisation to whole-mount embryos was performed according to Tautz and Pfeifle [69] with slight modifications [70].
Drosophila melanogaster stocks
The following stocks were used: bynapro (enhancer-trap line) [10] and yw67c23. The latter was used as the wild-type stock in this study. Stocks were raised on standard medium at 25 °C.
Antibody production
To generate anti-Otp antibodies against the Otp C-terminus a 0.6 kb fragment of our otp cDNA cW26/1 (corresponding to otp-RC) was amplified using the primer 10036–13 (5′-GGATCCACCAATGTCTTCCGCACCC-3′) to add a BamHI site (underlined) and the primer 10036–14 (5′-GAATTCGAATTGTAGTGTTCGTAGTTGTGTGG-3′) with an otp EcoRI site (underlined). The fragment was subcloned into the vector pCRII-TOPO (Invitrogen, Carlsbad, California, USA), cut with BamHI and EcoRI as a 577 bp fragment (position 886–1463 of CG10036-RC, Flybase) and cloned in frame into the pGEX-4 T1 expression vector (Amersham, Buckinghamshire, United Kingdom). The fusion protein of glutathione-S-transferase and Otp was purified as described [71]. Immunisation of guinea pigs was done by Pineda Antibody Service (Berlin, Germany).
Immunostaining
Embryos were collected, dechorionated with 50% bleach for 2 min, washed with 0.1% NaCl / 0.1% Triton X-100 and fixed for 12 min in 3.7% formaldehyde in PEM (100 mM PIPES, 1 mM EGTA, 1 mM MgCl2) and heptane. After removal of both phases, embryos were devitelinised in equal volumes of heptane and methanol by 2 min of vigorous shaking and washed three times with methanol. The 3rd instar larvae and adult hindguts were dissected in 1x phosphate buffered saline (PBS), fixed for 60 min in 2% paraformaldehyde in PBL and washed three times with 1x PBS containing 0.2% Triton X-100 (PBX) and then incubated for 3 × 5 min in methanol. Fixed embryos or larvae were washed 3 × 30 min and 6 × 30 min in PBX and blocked for 30 min in 5% normal horse serum and 10% PBX in PBS. Incubations with primary antibodies were done overnight at 4 °C. Samples were washed 3x5min and 6 × 30 min in PBX and blocked for 30 min in 5% normal horse serum and 10% PBX in PBS. After an overnight incubation with secondary antibodies at 4 °C embryos or larvae were washed 3 × 30 min and 6 x 30 min in PBX and mounted in Vectashield (Vector Laboratories). Images were obtained using an Olympus BX61 microscope (Olympus, Hamburg, Germany) for bright field and DIC microscopy or a Leica TCS SP5 microscope (Leica, Wetzlar, Germany) and a ZEISS LSM 710 microscope (Carl Zeiss AG, Oberkochen, Germany) for laser confocal microscopy. Images were processed using FIJI and ImageJ (NIH. Md., USA), Adobe Photoshop and Adobe Illustrator (Adobe Systems, San Jose, CA, USA).
The primary antibodies used were: rabbit anti-β-Galactosidase antibody (1:1000, Cappel), mouse anti-Crumbs antibody Cq4 (1:5, [44], DSHB), rabbit anti-Cleaved Drosophila Dcp-1 antibody (1:100, Cell Signaling), guinea pig anti-Orthopedia (1:400, this work). Secondary antibodies were goat anti-mouse, anti-rabbit and anti-guinea pig, conjugated with Alexa 488 or 568 (1:1000, Molecular Probes, Eugene, Oregon, USA).
Generation of an otp mutant by ends-out homologous recombination
The allele otpGT was generated as previously described [42]. The primers OtpGT3 (5′-GGCGCGCCTCAAAATAGGGCTTAAAACA-3′) and otpGT4 (5′-GGCGCGCCATAAACAGAATGCGTGCCAG-3′) were used in a PCR with BACR10P11 DNA as the template to amplify a 3.5 kb fragment (Otp-I) with AscI sites on both ends (underlined). The fragment was cut with AscI and cloned into the AscI site of the pW25 targeting vector (DGRC) to generate pW25/OtpI. Next, primers OtpGT1 (5′-GCGGCCGCATCGTAGTTTGCCAACTCGAGG-3′) and OtpGT2 (5′-GGTACCGCCGGATATGTGCAGGCGTG-3′) were used in another PCR with BACR10P11 DNA to add NotI and Acc65I sites (underlined) to a 3.7 kb fragment (OtpII), which was then cutted with NotI and Acc65I and cloned into the corresponding sites of the vector pW25/OtpI to generate pW25/OtpI+II. Four independent lines were obtained by germline transformation with the pW25/OtpI+II donor construct using standard techniques [72]. A donor target strain with the construct on the X-chromosome was crossed to yw; 70FLP, 70I-SceI, Sco/CyO flies and the F1 progenies were heat-shocked at 37 °C for 1 h on days 3, 4 and 5 after laying eggs. Among 106 crosses we identified red eyed flies in two crosses. One fly line, otpGT was balanced and the correct gene targeting event was verified by PCR amplification with genomic DNA extracted from otpGT flies as template and primers P1 (otp; 5′-CGTCCGGCACTTTGGCACG-3′) and P2 (pW25 vector; 5′-CGTGCTCATCGCGAGTACG-3′) or P3 (pW25 vector; 5′-GAGTGCCGTTTACTGTGCG-3′) and P4 (otp; 5′-GAGCAGCCCAGATTCCATGC-3′).
Amplification and cloning of the coding region of otp alleles
Amplifications of the coding region including the splice sites were performed using genomic DNA from fly stocks otp1024 and otp13064. For polymerase chain reactions Taq Polymerase from ThermoFisher Scientific (Waltham, Massachusetts, USA) was used according to supplier’s instructions. PCR products were sequenced by Starseq (Mainz, Germany). The region showing a sequence alteration in otp1024 DNA compared with the wild-type sequence was again PCR-amplified using more closely located primers. For otp1024 DNA the primers 1024–5 (5′-CTACCTTTAAATCTGTGGGTTTTGATGC-3′) and 1024–6 (5′-CATGCCAGGCGAAAAAGTGTC-3′) were used. The PCR product was subcloned into the TOPO vector pCR2.1 (ThermoFisher Scientific, Waltham, Massachusetts, USA), and at least 10 individual clones from PCR product cloning were checked by sequencing. Since the otp1024 DNA was generated from heterozygous flies approximately 50% of the clones showed the wild-type sequence and 50% the altered sequence due to the point mutation.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. Materials are available from the corresponding author on reasonable request.
Abbreviations
- BDPD:
-
Berkeley Drosophila Genome Project
- cDNA:
-
complementary desoxyribonucleic acid
- DSHB:
-
Developmental Studies Hybridoma Bank
- EST:
-
expressed sequence tag
References
Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. 2nd ed. Berlin: Springer; 1997.
Kusch T, Reuter R. Functions for Drosophila brachyenteron and forkhead in mesoderm specification and cell signalling. Development. 1999;126:3991–4003.
Teppas U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Dev Biol. 1994;161:563–96.
Harbecke R, Janning W. The segmentation gene Krüppel of Drosophila melanogaster has homeotic properties. Genes Dev. 1989;3:114–22.
Iwaki DD, Johansen KA, Singer JB, Lengyel JA. drumstick, bowl, and lines are required for pattering and cell rearrangement in the Drosophila embryonic hindgut. Dev Biol. 2001;240:611–26.
Smith AV, Orr-Weaver TL. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development. 1991;112:997–1008.
Fuß B, Meissner T, Bauer R, Lehmann C, Eckardt F, Hoch M. Control of endoreduplication domains in the Drosophila gut by the knirps and knirps-related genes. Mech Dev. 2001;100:15–23.
Takashima S, Murakami R. Regulation of pattern formation in the Drosophila hindgut by wg, hh, dpp and en. Mech Dev. 2001;101:79–90.
Hoch M, Pankratz MJ. Control of gut development by fork head and cell signaling molecules in Drosophila. Mech Dev. 1996;58:3–14.
Murakami R, Shigenaga A, Kawakita M, Takimoto K, Yamaoka I, Akasaka K, Shimada H. aproctous, a locus that is necessary for the development of the proctodeum in Drosophila embryos, encodes a homolog of the vertebrate Brachyury gene. Roux Arch Dev Biol. 1995;205:89–96.
Lengyel JA, Iwaki DD. It takes guts: the Drosophila hindgut as a model system for organogenesis. Dev Bio. 2002;243:1–19.
Strecker TR, Kongsuwan K, Lengyel JA, Merriam JR. The zygotic mutant tailless affects the anterior and posterior ectodermal regions of the Drosophila embryo. Dev Biol. 1986;113:64–76.
Kispert A, Herrmann BG, Leptin M, Reuter R. Homologs of mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev. 1994;8:2137–50.
Weigel D, Jürgens G, Küttner F, Seifert E, Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–58.
Wang L, Coulter DE. bowel, an odd-skipped homolog, functions in the terminal pathway during Drosophila embryogenesis. EMBO J. 1996;15:3182–96.
Baker NE. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J. 1987;6:1765–73.
Mlodzik M, Fjose A, Gehring WJ. Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 1985;4:2961–9.
Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–63.
Harbecke R, Lengyel JA. Genes controlling posterior gut development in the Drosophila embryo. Roux Arch Dev Biol. 1995;204:308–29.
Skaer H, Martinez AA. The wingless product is required for cell proliferation in the Malpighian tubule anlage of Drosophila melanogaster. Development. 1992;116:745–54.
Wu LH, Lengyel JA. Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development. 1998;125:2433–42.
Singer JB, Harbecke R, Kusch T, Reuter R, Lengyel JA. Drosophila brachyenteron regulates gene activity and morphogenesis in the gut. Development. 1996;122:3707–18.
Jack J, Myette G. The genes raw and ribbon are required for proper shape of tubular epithelial tissues in Drosophila. Genetics. 1997;147:243–53.
Blake KJ, Myette G, Jack J. The products of ribbon and raw are necessary for proper cell shape and cellular localization of nonmuscle myosin in Drosophila. Dev Biol. 2003;1998:177–88.
Liu X, Kiss I, Lengyel JA. Identification of genes controlling Malpighian tubule and other epithelial morphogenesis in Drosophila melanogaster. Genetics. 1999;151:685–95.
Sánchez-Soriano N, Russel S. Regulatory mutations of the Drosophila sox gene Dichaete reveal new functions in embryonic brain and hindgut development. Dev Biol. 2000;220:307–21.
Simeone A, D’Apice MR, Nigro V, Casanova J, Graziani F, Acampora D, Avantaggiato V. Orthopedia, a novel homeobox-containing gene expressed in the developing CNS of both mouse and Drosophila. Neuron. 1994;13:83–101.
Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–7.
Eggert T, Hauck B, Hildebrandt N, Gehring WJ, Walldorf U. Isolation of a Drosophila homolog of the vertebrate homeobox gene Rx and its possible role in brain and eye development. Proc Natl Acad Sci U S A. 1998;95:2343–8.
Walldorf U, Kiewe A, Wickert M, Ronshaugen M, McGinnis W. Homeobrain, a novel paired-like homeobox gene is expressed in the Drosophila brain. Mech Dev. 2000;96:141–4.
Kusch T, Storck T, Walldorf U, Reuter R. Brachyury proteins regulate target genes through modular binding sites in a cooperative fashion. Genes Dev. 2002;16:518–29.
Nederbragt AJ, Welscher P, van den Driesche S, van Loon AE, Dictus WJ. Novel and conserved roles for orthodenticle/otx and orthopedia/otp orthologs in the gastropod mollusc Patella vulgata. Dev Genes Evol. 2002;212:330–7.
Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129:1389–400.
Del Giacco L, Sordino P, Pistocchi A, Andreakis N, Tarallo R, Di Benedetto B, Cotelli F. Differential regulation of the zebrafish orthopedia I gene during fate determination of diencephalic neurons. BMC Dev Biol. 2006;6:50.
Caqueret A, Coumailleau P, Michaud JL. Regionalization of the anterior hypothalamus in the chick embryo. Dev Dyn. 2005;233:652–8.
Lin X, State MW, Vaccarino FM, Greally J, Hass M, Lackman JF. Identification, chromosomal assignment, and expression analysis of the human homeodomain-containing gene Orthopedia (OTP). Genomics. 1999;60:96–104.
Del Giacco L, Pistocchi A, Cotelli F, Fortunato AE, Sordino P. A peek inside the neurosecretory brain through Orthopedia lenses. Dev Dyn. 2008;237:2295–303.
Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR. And the FlyBase consortium. FlyBase 2.0: the next generation. Nucl Acids Res. 2019;47:D759–65.
Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature. 2008;454:651–5.
Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–8.
Rong YS, Golic KG. A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–12.
Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–61.
Grigliati T. In: Rickwood D, Hames BD, editors. Drosophila - A Practical Approach. Oxford: IRL Press; 1986. p. 39–58.
Tepass U, Theres C, Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–99.
Gregory SL, Kortschak RD, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA binding proteins. Mol Cell Biol. 1996;16:792–9.
Shandala T, Kortschak RD, Gregory S, Saint R. The Drosophila dead ringer gene is required for early embryonic patterning through regulation of argos and buttonhead expression. Development. 1999;126:4341–9.
White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–83.
Florentin A, Arama E. Caspase levels and execution efficiencies determine the apoptotic potential of the cell. J Cell Biol. 2012;196:513–27.
Green RB, Hatini V, Johansen KA, Liu X-J, Lengyel JA. Drumstick is a zinc finger protein that antagonizes lines to control patterning and morphogenesis of the Drosophila hindgut. Development. 2002;129:3645–56.
Fox DT, Spradling AC. The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell. 2009;5:290–7.
Takashima S, Paul M, Aghajanian P, Younossi-Hartenstein A, Hartenstein V. Migration of Drosophila intestinal stem cells across organ boundaries. Development. 2013;140:1903–11. https://doi.org/10.1242/dev.082933.
Messiaen L, Wimmer K. NF1 mutational spectrum. In: Kaufmann D, editor. Neurofibromatoses, vol. 16. Basel: Karger; 2008. p. 63–77.
Sanz DJ, Hollywood JA, Scallan MF, Harrison PT. Cas9/gRNA targeted excision of cystic fibrosis-causing deep-intronic splicing mutations restores normal splicing of CFTR mRNA. PLoS One. 2017;12:e0184009.
Abramowicz A, Gos M. Splicing mutations in human genetic disorders: examples, detection, and confirmation. J Appl Genetics. 2018;59:253–68.
Carthew RW, Agbu P, Giri R. MicroRNA function in Drosophila melanogaster. Semin Cell Dev Biol. 2017;65:29–37.
Eaton JL, Holmqvist B, Glasgow E. Ontogeny of vasotocin-expressing cells in zebrafish: selective requirement for the transcriptional regulators orthopedia and single-minded 1 in the preoptic area. Dev Dyn. 2008;237:995–1005.
Filippi A, Jainok C, Driever W. Analysis of transcriptional codes for zebrafish dopaminergic neurons reveals essential functions of Arx and Isl1 in prethalamic dopaminergic neuron development. Dev Biol. 2012;369:133–49.
Fernandes AM, Beddows E, Filippi A, Driever W. Orthopedia transcription factor otpa and otpb paralogous genes function during dopaminergic and neuroendocrine cell specification in larval zebrafish. PLoS One. 2013;8:e75002.
Amir-Zilberstein L, Blechmann J, Sztainberg Y, Norton WHJ, Reuveny A, Borodovsky N, Tahor M, Bonkowsky JL, Bally-Cuif L, Chen A, Levkowitz G. Homeodomain protein Otp and activity-dependent splicing modulate neuronal adaptation to stress. Neuron. 2012;73:279–91.
Gutierrez-Triana JA, Herget U, Lichtner P, Castillo-Ramirez LA, Ryu S. A vertebrate-conserved cis-regulatory module for targeted expression in the main hypothalamic regulatory region for the stress response. BMC Dev Biol. 2014;14:41.
Wircer E, Blechman J, Borodovsky N, Tsoory M, Nunes AR, Oliveira RF, Levkovitz G. Homeodomain protein Otp affects developmental neuropeptide switching in oxytocin neurons associated with the long-term effect on social behavior. eLife. 2017;6:e22170.
Acampora D, Postiglione MP, Avantaggiato V, Di Bonito M, Vaccarino FM, Michaud J, Simeone A. Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the Orthopedia gene. Genes Dev. 1999;13:2787–800.
Wang W, Lufkin T. The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev Biol. 2000;227:432–49.
Moir L, Bochukova EG, Dumbell R, Banks G, Bains RS, et al. Disruption of the homeodomain transcription factor orthopedia homeobox (Otp) is associated with obesity and anxiety. Mol Met. 2017;6:1419–28.
Caplin ME, Baudin E, Ferolla P, Filosso P, Garcia-Yuste M, Lim E, Oberg K, Pelosi G, Perren A, Rossi RE, Travis WD. Pulmonary neuroendocrine (carcinoid) tumors: European neuroendocrine tumor society expert consensus and recommendations for best practice for typical and atypical pulmonary carcinoids. Ann Oncol. 2015;26:1604–20.
Swarts DR, Van Neste L, Henfling ME, Eijkenboom I, Eijk PP, van Velthuysen ML, Vink A, Volante M, Ylstra B, Van Criekinge W, van Engeland M, Ramaekers FC, Speel EJ. An exploration of pathways involved in lung carcinoid progression using gene expression profiling. Carcinogenesis. 2013;34:2726–37.
Swarts DR, Henfling ME, Van Neste L, van Suylen RJ, Dingemans AM, Dinjens WN, Haesevoets A, Rudelius M, Thunnissen E, Volante M, Van Criekinge W, van Engeland M, Ramaekers FC, Speel EJ. CD44 and OTP are strong prognostic markers for pulmonary carcinoids. Clin Cancer Res. 2013;19:2197–207.
Mohler J, Vani K. Molecular organization and embryonic expression of the hedgehog gene involved in cell-cell communication in segmental patterning of Drosophila. Development. 1992;115:957–71.
Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–5.
Walldorf U, Gehring WJ. Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. EMBO J. 1992;11:2247–59.
Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40.
Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–53.
Acknowledgements
We are grateful to Rolf Reuter, the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank for fly stocks and antibodies. We thank Michael Faust for his support, Doris Jann and Sophia Streit for their technical assistance and Dieter Bruns and Peter Lipp for using their confocal microscopes. We thank the two anonymous reviewers for their comments and suggestions which improved the manuscript.
Funding
Not applicable. Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
KH, NB, DK, and UW conceived, designed the experiments, and analysed the data. UW wrote the manuscript, and all authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declares that there are no competing financial, personal, or professional interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hildebrandt, K., Bach, N., Kolb, D. et al. The homeodomain transcription factor Orthopedia is involved in development of the Drosophila hindgut. Hereditas 157, 46 (2020). https://doi.org/10.1186/s41065-020-00160-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41065-020-00160-y