Abstract
Purpose
We aimed to conduct a meta-analysis to accurately evaluate the potential association between ADIPOQ rs2241766 gene SNP and breast cancer risk.
Methods
A systematic literature search on Cochrane Library, PubMed, Embase, Web of Science and China National Knowledge Infrastructure (CNKI) identified 8 articles with 1692 cases and 1890 controls. Strength of association was evaluated by pooled odds ratio (OR), 95 % confidence interval (CI) and p value. Funnel plots and Begger’s regression test were applied for testing the publication bias. Statistical analysis of all data was performed by Stata 12.0.
Results
The meta-analysis results indicated that the ADIPOQ rs2241766 gene polymorphism did not significantly associated with the risk of breast cancer for these genetic models (TT vs. TG + GG: OR = 1.20, 95 % CI = 0.77–1.89, p=0.417; TT + TG vs. GG: OR = 1.05, 95 % CI = 0.71–1.56, p=0.805; T vs. G: OR =1.17, 95 % CI = 0.79–1.74, p=0.437).
Conclusions
This study indicated that no significant relationship between the ADIPOQ rs2241766 SNP and breast cancer. Further large-scale and well-designed studies will be indispensable to confirm our result.
Similar content being viewed by others
Introduction
Breast cancer was the most common malignant tumor in women, which was the second most common malignant tumor worldwide [1]. According to data from the World Health Organization and the International Agency for Research on Cancer in 2012, a total of 1.67 million breast cancer patients were confirmed worldwide. This number accounted for 11.8 % of all tumors [2]. So far, the pathogenesis of breast cancer remains unclear.
Recent studies had shown that adiponectin (ADIPOQ) was inversely related to breast cancer and other tumors [3,4,5,6]. ADIPOQ gene was found on the 3q27 chromosomal expressed by adipose tissue and had more than 620 variants [7, 8]. ADIPOQ gene polymorphism was closely related to cancer risk by influencing plasma level of ADIPOQ [9]. To date, previous studies on the relationship between the polymorphism of ADIPOQ rs2241766 gene and breast cancer susceptibility were limited and rather contradictory [10,11,12,13,14,15,16,17]. For lack of powerful evidence to provide a reliable conclusion in a single study, we conducted a comprehensive meta-analysis to assess the strength relationship between ADIPOQ rs2241766 gene polymorphism and breast cancer risk, which would have much greater possibility to reach reasonably reliable conclusions.
Materials and methods
Publication search
We systematically searched on database of Cochrane Library, Pubmed, Embase, Web of Science and China National Knowledge Infrastructure, up to October 31, 2019, using the following terms: (“ADIPOQ” OR “adiponectin receptor” OR “rs2241766”) AND (“variant” OR “polymorphism” OR “mutation”) AND (“breast cancer” OR “breast tumor”). Two investigators manually checked the reference of retrieved articles and extracted the publications independently. In addition, only English and Chinese articles were included.
Selection criteria
All selected studies complied with the inclusion criteria: (1) full text can be found; (2) case-control studies focused on relationship between the ADIPOQ rs2241766 polymorphism and risk of breast cancer; (3) ADIPOQ rs2241766 genotype was obtained. Main exclusion criteria as followed: (1) repeat of other articles; (2) not case–control studies; (3) unpublished studies, conference articles, meta-analysis and systematic evaluations; (4) pedigree studies. Consulting the Preferred Reporting Project (PRISMA) Guide for Systematic Evaluation and Meta-Analysis[18], by screening all retrieved literatures, we constructed an information flow diagram about the final eligible data.
Data extraction
Two investigators extracted data independently according to the selection criteria. The following items were collected: first author, country, publication year, amount of cases and controls, Hardy-Weinberg equilibrium, control group source and the availability of ADIPOQ rs2241766 genotype. Only the article with maximum sample size was selected while same data appearing in multiple publications. To insure the data accuracy, a third investigator reviewed the final results. Discussions were executed to solve disagreements.
Study quality assessment
Two researchers performed independent quality assessment for each eligible article according to 9-point Newcastle-Ottawa Scale (NOS) applied to quality evaluation of observational studies [19]. Different results from two evaluators were solved by the third assessor. Assessment score principally included these aspects: (1) case and control selection (4 point); (2) Confounding factor quality corrected in cases and controls (2 point); (3) exposure ascertainment (3 point). The total scores ranged from 0 to 9, and scores above 6 indicate high quality.
Statistical analysis
Odds ratio (OR) and 95 % confidence interval (CI) were calculated to estimate the relationship between ADIPOQ rs2241766 gene SNP and the risk of breast cancer. The Chi-square based Q-test and I-squared test was used to analyze the heterogeneity (P<0.10 suggested Heterogeneity) [20, 21]. The pooled OR was estimated by fixed effect model (Mantel–Haenszel) when no heterogeneity existed. Otherwise, the pooled OR was estimated by random effect model (DerSimonian and Laird) [22, 23]. In controls, Chi-square test was used to examine Hardy–Weinberg equilibrium (HWE). In order to estimate the influence of the pooled ORs caused by individual data set, we performed sensitivity analysis for each comparison models respectively. The publication bias was tested by Funnel plot and Begg linear regression [24, 25]. Stata 12.0 was used to perform all analysis (Stata Corp, College Station, United States).
Results
Characteristic of studies
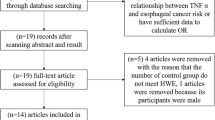
Flow diagram for the retrieve strategy was demonstrated in Fig. 1. 311 publications were identified initially. 68 duplicate publications were excluded by verified and deleted, while 243 publications entered our study. 54 publications were reviewed for full-text review by reading title and abstract. Ultimately, 8 studies with 1692 breast cancer patients and 1890 controls were screened out for the final meta-analysis which published between 2008 and 2019. The gene distributions in control groups were all consistent with HWE. In addition, all studies were high quality because of the NOS scores ranging from 7 to 8. The relevant feature information was presented in Table 1.
Meta-analysis results
Chi-square was used to determine the heterogeneity of the three genetic models. The heterogeneityre was significant in these models according to Table 2 (TT vs. TG + GG (recessive model): p<0.001, I2 = 84.2 %; T vs. G (allele model): p<0.001, I2= 84.3 %). However, the heterogeneity was not considered significant in these models (TT + TG vs. GG (dominant model): p=0.308, I2 = 16.0 %) Fig. 2. So the recessive model and allele models were analyzed by random effect model. The fixed was applied to analyze the dominant model. There was no significant association between ADIPOQ rs2241766 polymorphism and breast cancer risk under the model of TT vs. TG + GG (OR = 1.20, 95 % CI = 0.77–1.89, p=0.417), TT + TG vs. GG (OR =1.05, 95 % CI = 0.71–1.56, p=0.805), C vs. T (OR =1.17, 95 % CI = 0.79–1.74, p=0.437). We also performed a subgroup analysis according to ethnicity, source of control, case size and control size. We find the statistically significant association for the dominant model which observed in Asian population (OR=1.48, 95 %CI=1.08-2.05), control from population (OR=1.49, 95 %CI=1.01-2.18) and studies with less or more than 100 controls (OR=1.67, 95 %CI=1.01-2.77), and less or more than 100 cases. The pooled OR and 95 % CI of the relationship between ADIPOQ rs2241766 polymorphism and breast cancer were in Table 1. A sensitivity analysis was conducted to reflect the effect of each independent study on the whole studies. As shown in Fig. 3, none of the study affected the overall results of. Begg’s funnel plot was used to assess the publication bias. The results showed that there was no publication bias reflected in three genetic models (TT vs. TG + GG: p=0.458; TT + TG vs. GG: p=0.881; T vs. G: p=0.322) (Fig. 4).
Discussion
In recent years, several case–control studies have shown an association between the importance of ADIPOQ rs2241766 gene SNP and breast cancer risk, however, to date, it is still inconclusion. The first study had been reported ADIPOQ rs2241766 gene polymorphisms in relation to breast cancer risk in 2008 [10], in this study, the association between the high expression of rs2241766 G allele (GT and GG genotype) and low risk of breast cancer was detected. In 2019, Macias-Gomez et al. [16] found that the G allele of ADIPOQ rs2241766 polymorphisms and the TG+GG genotype had protective effects on the occurrence of breast cancer. Series of studies supported the conclusion while others objected. Pasha et al. [17] detected that the ADIPOQ rs2241766 SNP might cause breast cancer (OR=6.2, 95 %CI=1.3–29.6). Al Khaldi et al. [12] found that individuals with GG genotypes showed an increased risk of breast cancer (OR =2.1, 95 %CI=1.1–4.1). Others studies suggested that there was no association between ADIPOQ rs2241766 gene SNP and increased risk of breast cancer [11, 13,14,15].
In the recessive model (OR = 1.20, 95 %CI = 0.77–1.89), dominant model (OR = 1.05, 95 %CI = 0.71–1.56) and allele model (OR = 1.17, 95 %CI = 0.79–1.74), the ADIPOQ rs2241766 polymorphism was not significantly associated with breast cancer risk. Considerable heterogeneity was discovered among these studies. We also conducted a subgroup analysis according to racial classification, case size, control size and source of control. In dominant genetic model, the significant correlation was not found in stratification analysis by racial classification, case size, control size and source of control. But in Asians, the statistically significant association for the dominant model was observed (OR=1.48, 95 %CI=1.08-2.05), control from population (OR=1.49, 95 %CI=1.01-2.18) and studies with less than 100 controls (OR=1.67, 95 %CI=1.01-2.77). Moreover, the significant associations were observed in allele genetic model in stratified analysis among Asians (OR=1.36, 95 %CI=1.04-1.78) and studies with less than 100 controls (OR=1.52, 95 %CI=1.00-2.30). Because of the limited samples and finite studies [11, 12], it would be immature to conclude that the ADIPOQ rs2241766 SNP has no relationship with breast cancer. New further discoveries will appear and the information will be constantly renovated. There should be plenty of research data to support more objective results.
The detailed mechanism of the association between ADIPOQ rs2241766 and breast cancer is unclear. Previous epidemiological studies [26, 27] have confirmed a significant association between obesity and some adipokines and breast cancer risk. Logically, adiponectin may play a role in the development of breast cancer. Uncontrolled cellular proliferation is a hallmark of tumorigenesis. Adiponectin is an important regulator of cell proliferation and apoptosis. Adiponectin has been shown to significantly suppress the proliferation of MDAMB-231 cells by arresting the cells at G0/G1 phase and inducing apoptosis [28]. Furthermore, adiponectin also significantly inhibits cell proliferation induced by leptin, oxidized, low-density lipoprotein, platelet-derived growth factor BB, basic fibroblast growth factor (bFGF), and heparinbinding epidermal growth factor-like growth factor [28].
Several limitations in the study should be considered. First, only 8 studies were selected into our meta-analysis. The sample size and amount of studies were comparatively small, which affected the reliability of the results. Second, the case size and control size of some studies were relatively small to confirm the risk of ADIPOQ rs2241766 SNP. Finally, in all genetic models, the heterogeneity of the recessive model and allele model may have a significant impact on the result of meta-analysis.
Conclusions
In conclusion, we find the significant associations in the recessive model and allele genetic model in stratification analysis. The risk of ADIPOQ rs2241766 SNP could not be confirmed due to the relatively limited sample and small amount of study. Scholars had conducted extensive research on the susceptibility factors of breast cancer, which believed that the breast cancer occurrence was influenced by multiple factors [29,30,31,32,33], especially genetic factors and life-style. Therefore, the influence of confounding risk factors will be eliminated by further studies, such as age, BMI and environmental factors.
Availability of data and materials
Not applicable.
Abbreviations
- CNKI:
-
China National Knowledge Infrastructure
- ADIPOQ:
-
adiponectin
- IARC:
-
International Agency for Research on Cancer
- WHO:
-
World Health Organization
- NOS:
-
Newcastle-Ottawa Scale
- OR:
-
odds ratio
- CI:
-
confidence interval
References
da Costa Vieira RA, Biller G, Uemura G, Ruiz CA, Curado MP. Breast cancer screening in developing countries. Clinics. 2017;72(4):244–53.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136(5):359–86.
Gu CY, Li QX, Zhu Y, Wang MY, Shi TY, Yang YY, Wang JC, Jin L, Wei QY, Ye DW. Genetic variations of the ADIPOQgene and risk of prostate cancer in Chinese Han men. Asian journal of andrology. 2014;16(6):878–83.
Parida S, Siddharth S, Sharma D. Adiponectin, Obesity, and Cancer: Clash of the Bigwigs in Health and Disease. International journal of molecular sciences 2019; 20(10).
Sonmez B, Seker M, Bilici A, Yavuz Erkal F, Oven Ustaalioglu BB, Gumus M, Ozturk Guler D, Karaduman M, Gezen C, Eser M, et al. Is there any correlation among adiponectin levels in serum, tumor tissue and normal tissue of the same patients wih breast cancer? Journal of BUON: official journal of the Balkan Union of Oncology. 2011;16(2):227–32.
Zhang G, Gu C, Zhu Y, Luo L, Dong D, Wan F, Zhang H, Shi G, Sun L, Ye D. ADIPOQ polymorphism rs182052 is associated with clear cell renal cell carcinoma. Cancer Sci. 2015;106(6):687–91.
Takahashi M, Arita Y, Yamagata K, Matsukawa Y, Okutomi K, Horie M, Shimomura I, Hotta K, Kuriyama H, Kihara S, et al. Genomic structure and mutations in adipose-specific gene, adiponectin. International journal of obesity related metabolic disorders: journal of the International Association for the Study of Obesity. 2000;24(7):861–8.
Chandran M, Phillips SA, Ciaraldi T, Henry RR. Adiponectin: more than just another fat cell hormone? Diabetes Care. 2003;26(8):2442–50.
Vona-Davis L, Howard-McNatt M, Rose DP. Adiposity, type 2 diabetes and the metabolic syndrome in breast cancer. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2007;8(5):395–408.
Kaklamani VG, Sadim M, Hsi A, Offit K, Oddoux C, Ostrer H, Ahsan H, Pasche B, Mantzoros C. Variants of the adiponectin and adiponectin receptor 1 genes and breast cancer risk. Cancer research. 2008;68(9):3178–84.
Hongxia Cui. Association of adiponectin and adiponectin gene polymorphisms with risk of breast cancer. Shanxi Medical University; 2009.
Al Khaldi RM, Al Mulla F, Al Awadhi S, Kapila K, Mojiminiyi OA. Associations of single nucleotide polymorphisms in the adiponectin gene with adiponectin levels and cardio-metabolic risk factors in patients with cancer. Disease markers. 2011;30(4):197–212.
Kaklamani VG, Hoffmann TJ, Thornton TA, Hayes G, Chlebowski R, Van Horn L, Mantzoros C. Adiponectin pathway polymorphisms and risk of breast cancer in African Americans and Hispanics in the Women’s Health Initiative. Breast cancer research treatment. 2013;139(2):461–8.
Erbay B, Yilmaz TU, Eraldemir C, Uren N, Tiryaki C, Ergul E, Utkan Z. The Relationship between Adiponectin and Breast Cancer. The journal of breast health. 2016;12(2):67–71.
Khandouzi M, Deka M, Baruah MN, Rozati R, Kordafshari M, Kashyap P, Kumari N. Genetic variation in adiponectin, leptin and leptin receptors with reference to risk of breast cancer in Northeast obese women in India. Bioscience Biotechnology Research Communications. 2016;9(2):163–70.
Macias-Gomez NM, Hernandez-Terrones MC, Ramirez-Guerrero AA, Leal-Ugarte E, Gutierrez-Angulo M, Peregrina-Sandoval J. ADIPOQ rs2241766 SNP as protective marker against DIBC development in Mexican population. PloS one. 2019;14(3):e0214080.
Pasha HF, Mohamed RH, Toam MM, Yehia AM. Genetic and epigenetic modifications of adiponectin gene: Potential association with breast cancer risk. J Gene Med. 2019;21(10):e3120.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61(7):634–45.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
McKenzie JE, Beller EM, Forbes AB. Introduction to systematic reviews and meta-analysis. Respirology. 2016;21(4):626–37.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Maskarinec G, Woolcott C, Steude JS, Franke AA, Cooney RV. The relation of leptin and adiponectin with breast density among premenopausal women. Eur J Cancer Prev. 2010;19:55–60.
Tworoger SS, Eliassen AH, Kelesidis T, Colditz GA, Willett WC, Mantzoros CS, et al. Plasma adiponectin concentrations and risk of incident breast cancer. J Clin Endocrinol Metab. 2007;92:1510–6.
Chen X, Wang Y. Adiponectin and breast cancer. Med Oncol. 2011 Dec;28(4):1288–95.
Rojas KE, Matthews N, Raker C, Clark MA, Onstad M, Stuckey A, Gass J. Body mass index (BMI), postoperative appearance satisfaction, and sexual function in breast cancer survivorship. Journal of cancer survivorship: research practice. 2018;12(1):127–33.
Suzuki Y, Tsunoda H, Kimura T, Yamauchi H. BMI change and abdominal circumference are risk factors for breast cancer, even in Asian women. Breast cancer research treatment. 2017;166(3):919–25.
Li T, Tang L, Gandomkar Z, Heard R, Mello-Thoms C, Shao Z, Brennan P. Mammographic density and other risk factors for breast cancer among women in China. The breast journal. 2018;24(3):426–8.
Fu XJ, Shi XJ, Lin K, Lin H, Huang WH, Zhang GJ, Au WW. Environmental and DNA repair risk factors for breast cancer in South China. Int J Hyg Environ Health. 2015;218(3):313–8.
Hegewald J, Schubert M, Wagner M, Droge P, Prote U, Swart E, Mohler U, Zeeb H, Seidler A. Breast cancer and exposure to aircraft, road, and railway-noise: a case-control study based on health insurance records. Scand J Work Environ Health. 2017;43(6):509–18.
Acknowledgements
We appreciate the cooperation of the families and individuals who cooperated in this study.
Funding
China Postdoctoral Science Foundation Grant (No. 2019M651216).
Author information
Authors and Affiliations
Contributions
Manuscript writing, articles search, editing and review were conducted by Xue Hu and Wan Wang; Chunguo Cui and Tong Sun performed data analysis and evaluation the quality of the selected studies. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hu, X., Cui, C., Sun, T. et al. Associations between ADIPOQ rs2241766 SNP and breast cancer risk: a systematic review and a meta-analysis. Genes and Environ 43, 48 (2021). https://doi.org/10.1186/s41021-021-00221-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41021-021-00221-2